Ni(2+) transport and accumulation in Rhodospirillum rubrum
- PMID: 10419953
- PMCID: PMC103586
- DOI: 10.1128/JB.181.15.4554-4560.1999
Ni(2+) transport and accumulation in Rhodospirillum rubrum
Abstract
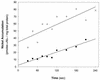
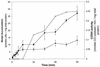
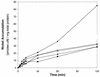
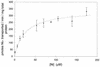
The cooCTJ gene products are coexpressed with CO-dehydrogenase (CODH) and facilitate in vivo nickel insertion into CODH. A Ni(2+) transport assay was used to monitor uptake and accumulation of (63)Ni(2+) into R. rubrum and to observe the effect of mutations in the cooC, cooT, and cooJ genes on (63)Ni(2+) transport and accumulation. Cells grown either in the presence or absence of CO transported Ni(2+) with a K(m) of 19 +/- 4 microM and a V(max) of 310 +/- 22 pmol of Ni/min/mg of total protein. Insertional mutations disrupting the reading frame of the cooCTJ genes, either individually or all three genes simultaneously, transported Ni(2+) the same as wild-type cells. The nickel specificity for transport was tested by conducting the transport assay in the presence of other divalent metal ions. At a 17-fold excess Mn(2+), Mg(2+), Ca(2+), and Zn(2+) showed no inhibition of (63)Ni(2+) transport but Co(2+), Cd(2+), and Cu(2+) inhibited transport 35, 58, and 66%, respectively. Nickel transport was inhibited by cold (50% at 4 degrees C), by protonophores (carbonyl cyanide m-chlorophenylhydrazone, 44%, and 2,4-dinitrophenol, 26%), by sodium azide (25%), and hydroxyl amine (33%). Inhibitors of ATP synthase (N, N'-dicyclohexylcarbodiimide and oligomycin) and incubation of cells in the dark stimulated Ni(2+) transport. (63)Ni accumulation after 2 h was four times greater in CO-induced cells than in cells not exposed to CO. The CO-stimulated (63)Ni(2+) accumulation coincided with the appearance of CODH activity in the culture, suggesting that the (63)Ni(2+) was accumulating in CODH. The cooC, cooT, and cooJ genes are required for the increased (63)Ni(2+) accumulation observed upon CO exposure because cells containing mutations disrupting any or all of these genes accumulated (63)Ni(2+) like cells unexposed to CO.
Figures
Similar articles
-
Maturation of the [Ni-4Fe-4S] active site of carbon monoxide dehydrogenases.J Biol Inorg Chem. 2018 Jun;23(4):613-620. doi: 10.1007/s00775-018-1541-0. Epub 2018 Feb 14. J Biol Inorg Chem. 2018. PMID: 29445873 Free PMC article. Review.
-
In vivo nickel insertion into the carbon monoxide dehydrogenase of Rhodospirillum rubrum: molecular and physiological characterization of cooCTJ.J Bacteriol. 1997 Apr;179(7):2259-66. doi: 10.1128/jb.179.7.2259-2266.1997. J Bacteriol. 1997. PMID: 9079911 Free PMC article.
-
Purification and characterization of membrane-associated CooC protein and its functional role in the insertion of nickel into carbon monoxide dehydrogenase from Rhodospirillum rubrum.J Biol Chem. 2001 Oct 19;276(42):38602-9. doi: 10.1074/jbc.M104945200. Epub 2001 Aug 15. J Biol Chem. 2001. PMID: 11507093
-
The identification, purification, and characterization of CooJ. A nickel-binding protein that is co-regulated with the Ni-containing CO dehydrogenase from Rhodospirillum rubrum.J Biol Chem. 1998 Apr 17;273(16):10019-25. doi: 10.1074/jbc.273.16.10019. J Biol Chem. 1998. PMID: 9545348
-
The metalloclusters of carbon monoxide dehydrogenase/acetyl-CoA synthase: a story in pictures.J Biol Inorg Chem. 2004 Jul;9(5):511-5. doi: 10.1007/s00775-004-0563-y. Epub 2004 Jun 18. J Biol Inorg Chem. 2004. PMID: 15221484 Review.
Cited by
-
The carbon monoxide dehydrogenase accessory protein CooJ is a histidine-rich multidomain dimer containing an unexpected Ni(II)-binding site.J Biol Chem. 2019 May 10;294(19):7601-7614. doi: 10.1074/jbc.RA119.008011. Epub 2019 Mar 11. J Biol Chem. 2019. PMID: 30858174 Free PMC article.
-
Maturation of the [Ni-4Fe-4S] active site of carbon monoxide dehydrogenases.J Biol Inorg Chem. 2018 Jun;23(4):613-620. doi: 10.1007/s00775-018-1541-0. Epub 2018 Feb 14. J Biol Inorg Chem. 2018. PMID: 29445873 Free PMC article. Review.
-
Genome annotation provides insight into carbon monoxide and hydrogen metabolism in Rubrivivax gelatinosus.PLoS One. 2014 Dec 5;9(12):e114551. doi: 10.1371/journal.pone.0114551. eCollection 2014. PLoS One. 2014. PMID: 25479613 Free PMC article.
-
New insights into the mechanism of nickel insertion into carbon monoxide dehydrogenase: analysis of Rhodospirillum rubrum carbon monoxide dehydrogenase variants with substituted ligands to the [Fe3S4] portion of the active-site C-cluster.J Biol Inorg Chem. 2005 Dec;10(8):903-12. doi: 10.1007/s00775-005-0043-z. Epub 2005 Nov 8. J Biol Inorg Chem. 2005. PMID: 16283394
-
Bioconversion of CO to formate by artificially designed carbon monoxide:formate oxidoreductase in hyperthermophilic archaea.Commun Biol. 2022 Jun 3;5(1):539. doi: 10.1038/s42003-022-03513-7. Commun Biol. 2022. PMID: 35660788 Free PMC article.
References
-
- Baltscheffsky M. Photosynthetic phosphorylation. In: Clayton R K, Sistrom W R, editors. The photosynthetic bacteria. New York, N.Y: Plenum Press, Inc.; 1978. pp. 595–613.
-
- Baudet C, Sprott G D, Patel G B. Adsorption and uptake of nickel in Methanothrix concilii. Arch Microbiol. 1988;150:338–342.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

