The structure of multiple polypeptide domains determines the signal recognition particle targeting requirement of Escherichia coli inner membrane proteins
- PMID: 10419954
- PMCID: PMC103587
- DOI: 10.1128/JB.181.15.4561-4567.1999
The structure of multiple polypeptide domains determines the signal recognition particle targeting requirement of Escherichia coli inner membrane proteins
Abstract
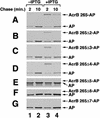
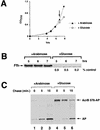
The signal recognition particle (SRP) targeting pathway is required for the efficient insertion of many polytopic inner membrane proteins (IMPs) into the Escherichia coli inner membrane, but in the absence of SRP protein export proceeds normally. To define the properties of IMPs that impose SRP dependence, we analyzed the targeting requirements of bitopic IMPs that are structurally intermediate between exported proteins and polytopic IMPs. We found that disruption of the SRP pathway inhibited the insertion of only a subset of bitopic IMPs. Studies on a model bitopic AcrB-alkaline phosphatase fusion protein (AcrB 265-AP) showed that the SRP requirement for efficient insertion correlated with the presence of a large periplasmic domain (P1). As previously reported, perturbation of the SRP pathway also affected the insertion of a polytopic AcrB-AP fusion. Even exhaustive SRP depletion, however, failed to block the insertion of any AcrB derivative by more than 50%. Taken together, these data suggest that many proteins that are normally targeted by SRP can utilize alternative targeting pathways and that the structure of both hydrophilic and membrane-spanning domains determines the degree to which the biogenesis of a protein is SRP dependent.
Figures



References
-
- Bernstein H D, Poritz M A, Strub K, Hoben P J, Brenner S, Walter P. Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature. 1989;340:482–486. - PubMed
-
- Brown S, Fournier M J. The 4.5S RNA gene of Escherichia coli is essential for cell growth. J Mol Biol. 1984;178:533–550. - PubMed
-
- Bukau B, Horwich A L. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. - PubMed
-
- Casadaban M J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;104:541–555. - PubMed
-
- Claros M G, von Heijne G. TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci. 1994;10:685–686. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

