MgATP binding and hydrolysis determinants of NtrC, a bacterial enhancer-binding protein
- PMID: 10419963
- PMCID: PMC103596
- DOI: 10.1128/JB.181.15.4628-4638.1999
MgATP binding and hydrolysis determinants of NtrC, a bacterial enhancer-binding protein
Abstract
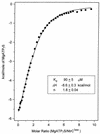
When phosphorylated, the dimeric form of nitrogen regulatory protein C (NtrC) of Salmonella typhimurium forms a larger oligomer(s) that can hydrolyze ATP and hence activate transcription by the sigma(54)-holoenzyme form of RNA polymerase. Studies of Mg-nucleoside triphosphate binding using a filter-binding assay indicated that phosphorylation is not required for nucleotide binding but probably controls nucleotide hydrolysis per se. Studies of binding by isothermal titration calorimetry indicated that the apparent K(d) of unphosphorylated NtrC for MgATPgammaS is 100 microM at 25 degrees C, and studies by filter binding indicated that the concentration of MgATP required for half-maximal binding is 130 microM at 37 degrees C. Filter-binding studies with mutant forms of NtrC defective in ATP hydrolysis implicated two regions of its central domain directly in nucleotide binding and three additional regions in hydrolysis. All five are highly conserved among activators of sigma(54)-holoenzyme. Regions implicated in binding are the Walker A motif and the region around residues G355 to R358, which may interact with the nucleotide base. Regions implicated in nucleotide hydrolysis are residues S207 and E208, which have been proposed to lie in a region analogous to the switch I effector region of p21(ras) and other purine nucleotide-binding proteins; residue R294, which may be a catalytic residue; and residue D239, which is the conserved aspartate in the putative Walker B motif. D239 appears to play a role in binding the divalent cation essential for nucleotide hydrolysis. Electron paramagnetic resonance analysis of Mn(2+) binding indicated that the central domain of NtrC does not bind divalent cation strongly in the absence of nucleotide.
Figures



References
-
- Abrahams J P, Leslie A G, Lutter R, Walker J E. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. - PubMed
-
- Al-Shawi M K, Pasonage D, Senior A E. Directed mutagenesis of the strongly conserved aspartate 242 in the β-subunit of Escherichia coli proton-ATPase. J Biol Chem. 1988;263:19633–19639. - PubMed
-
- Ausubel F A, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994.
-
- Berchtold H, Reshetnikova L, Reiser C O, Schirmer N K, Sprinzl M, Hilgenfeld R. Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature. 1993;365:126–132. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

