Genetic structure of natural populations of Escherichia coli in wild hosts on different continents
- PMID: 10427022
- PMCID: PMC91507
- DOI: 10.1128/AEM.65.8.3373-3385.1999
Genetic structure of natural populations of Escherichia coli in wild hosts on different continents
Abstract
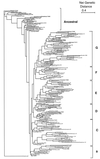
Current knowledge of genotypic and phenotypic diversity in the species Escherichia coli is based almost entirely on strains recovered from humans or zoo animals. In this study, we analyzed a collection of 202 strains obtained from 81 mammalian species representing 39 families and 14 orders in Australia and the Americas, as well as several reference strains; we also included a strain from a reptile and 10 from different families of birds collected in Mexico. The strains were characterized genotypically by multilocus enzyme electrophoresis (MLEE) and phenotypically by patterns of sugar utilization, antibiotic resistance, and plasmid profile. MLEE analysis yielded an estimated genetic diversity (H) of 0.682 for 11 loci. The observed genetic diversity in this sample is the greatest yet reported for E. coli. However, this genetic diversity is not randomly distributed; geographic effects and host taxonomic group accounted for most of the genetic differentiation. The genetic relationship among the strains showed that they are more associated by origin and host order than is expected by chance. In a dendrogram, the ancestral cluster includes primarily strains from Australia and ECOR strains from groups B and C. The most differentiated E. coli in our analysis are strains from Mexican carnivores and strains from humans, including those in the ECOR group A. The kinds and numbers of sugars utilized by the strains varied by host taxonomic group and country of origin. Strains isolated from bats were found to exploit the greatest range of sugars, while those from primates utilized the fewest. Toxins are more frequent in strains from rodents from both continents than in any other taxonomic group. Strains from Mexican wild mammals were, on average, as resistant to antibiotics as strains from humans in cities. On average, the Australian strains presented a lower antibiotic resistance than the Mexican strains. However, strains recovered from hosts in cities carried significantly more plasmids than did strains isolated from wild mammals. Previous studies have shown that natural populations of E. coli harbor an extensive genetic diversity that is organized in a limited number of clones. However, knowledge of this worldwide bacterium has been limited. Here, we suggest that the strains from a wide range of wild hosts from different regions of the world are organized in an ecotypic structure where adaptation to the host plays an important role in the population structure.
Figures
Comment in
-
Genetic diversity within E.coli.Appl Environ Microbiol. 2000 Nov;66(11):5104-5. doi: 10.1128/AEM.66.11.5104-5105.2000. Appl Environ Microbiol. 2000. PMID: 11184318 Free PMC article. No abstract available.
References
-
- Baquero F, Blazquez J. Evolution of antibiotic resistance. Trends Ecol Evol. 1998;12:482–488. - PubMed
-
- Blattner F R, Plunkett I G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. - PubMed
-
- Campbell I C. Island kingdom: Tonga ancient and modern. Christchurch, New Zealand: Canterbury University Press; 1992.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

