DAip1, a Dictyostelium homologue of the yeast actin-interacting protein 1, is involved in endocytosis, cytokinesis, and motility
- PMID: 10427097
- PMCID: PMC2156175
- DOI: 10.1083/jcb.146.2.453
DAip1, a Dictyostelium homologue of the yeast actin-interacting protein 1, is involved in endocytosis, cytokinesis, and motility
Abstract
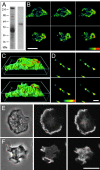
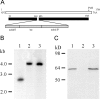
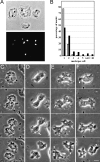
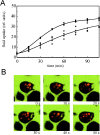
The 64-kD protein DAip1 from Dictyostelium contains nine WD40-repeats and is homologous to the actin-interacting protein 1, Aip1p, from Saccharomyces cerevisiae, and to related proteins from Caenorhabditis, Physarum, and higher eukaryotes. We show that DAip1 is localized to dynamic regions of the cell cortex that are enriched in filamentous actin: phagocytic cups, macropinosomes, lamellipodia, and other pseudopodia. In cells expressing green fluorescent protein (GFP)-tagged DAip1, the protein rapidly redistributes into newly formed cortical protrusions. Functions of DAip1 in vivo were assessed using null mutants generated by gene replacement, and by overexpressing DAip1. DAip1-null cells are impaired in growth and their rates of fluid-phase uptake, phagocytosis, and movement are reduced in comparison to wild-type rates. Cytokinesis is prolonged in DAip1-null cells and they tend to become multinucleate. On the basis of similar results obtained by DAip1 overexpression and effects of latrunculin-A treatment, we propose a function for DAip1 in the control of actin depolymerization in vivo, probably through interaction with cofilin. Our data suggest that DAip1 plays an important regulatory role in the rapid remodeling of the cortical actin meshwork.
Figures




Similar articles
-
Genetic evidence for concerted control of actin dynamics in cytokinesis, endocytic traffic, and cell motility by coronin and Aip1.Cytoskeleton (Hoboken). 2010 Jul;67(7):442-55. doi: 10.1002/cm.20456. Cytoskeleton (Hoboken). 2010. PMID: 20506401
-
Live dynamics of Dictyostelium cofilin suggests a role in remodeling actin latticework into bundles.J Cell Sci. 1997 Oct;110 ( Pt 19):2333-44. doi: 10.1242/jcs.110.19.2333. J Cell Sci. 1997. PMID: 9410873
-
Actin-depolymerizing factor and cofilin-1 play overlapping roles in promoting rapid F-actin depolymerization in mammalian nonmuscle cells.Mol Biol Cell. 2005 Feb;16(2):649-64. doi: 10.1091/mbc.e04-07-0555. Epub 2004 Nov 17. Mol Biol Cell. 2005. PMID: 15548599 Free PMC article.
-
The coronin family of actin-associated proteins.Trends Cell Biol. 1999 Sep;9(9):345-50. doi: 10.1016/s0962-8924(99)01620-7. Trends Cell Biol. 1999. PMID: 10461187 Review.
-
The actin cytoskeleton of Dictyostelium: a story told by mutants.J Cell Sci. 2000 Mar;113 ( Pt 5):759-66. doi: 10.1242/jcs.113.5.759. J Cell Sci. 2000. PMID: 10671366 Review.
Cited by
-
A Diaphanous-related formin links Ras signaling directly to actin assembly in macropinocytosis and phagocytosis.Proc Natl Acad Sci U S A. 2016 Nov 22;113(47):E7464-E7473. doi: 10.1073/pnas.1611024113. Epub 2016 Nov 7. Proc Natl Acad Sci U S A. 2016. PMID: 27821733 Free PMC article.
-
The localization of inner centromeric protein (INCENP) at the cleavage furrow is dependent on Kif12 and involves interactions of the N terminus of INCENP with the actin cytoskeleton.Mol Biol Cell. 2007 Sep;18(9):3366-74. doi: 10.1091/mbc.e06-10-0895. Epub 2007 Jun 13. Mol Biol Cell. 2007. PMID: 17567958 Free PMC article.
-
Self-organizing actin waves as planar phagocytic cup structures.Cell Adh Migr. 2009 Oct-Dec;3(4):373-82. doi: 10.4161/cam.3.4.9708. Epub 2009 Oct 1. Cell Adh Migr. 2009. PMID: 19855162 Free PMC article.
-
Small GTPases and phosphoinositides in the regulatory mechanisms of macropinosome formation and maturation.Front Physiol. 2014 Sep 30;5:374. doi: 10.3389/fphys.2014.00374. eCollection 2014. Front Physiol. 2014. PMID: 25324782 Free PMC article. Review.
-
Expression of actin-interacting protein 1 suppresses impaired chemotaxis of Dictyostelium cells lacking the Na+-H+ exchanger NHE1.Mol Biol Cell. 2010 Sep 15;21(18):3162-70. doi: 10.1091/mbc.E09-12-1058. Epub 2010 Jul 28. Mol Biol Cell. 2010. PMID: 20668166 Free PMC article.
References
-
- Adler H.J., Winnicki R.S., Gong T.-W.L., Lomax M.I. A gene upregulated in the acoustically damaged chick basilar papilla encodes a novel WD40 repeat protein. Genomics. 1999;56:59–69. - PubMed
-
- Aizawa H., Sutoh K., Tsubuki S., Kawashima S., Ishii A., Yahara I. Identification, characterization, and intracellular distribution of cofilin in Dictyostelium discoideum . J. Biol. Chem. 1995;270:10923–10932. - PubMed
-
- Aizawa H., Fukui Y., Yahara I. Live dynamics of Dictyostelium cofilin suggests a role in remodeling actin latticework into bundles. J. Cell Sci. 1997;110:2333–2344. - PubMed
-
- Aizawa H., Katadae M., Maruya M., Sameshima M., Murakami-Murofushi K., Yahara I. Contraction of actin bundles induced in Dictyostelium by overexpression of cofilin in response to hyperosmotic stress Mol. Biol. Cell. 9Suppl.1998. 93
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

