An aminoacyl-tRNA synthetase paralog with a catalytic role in histidine biosynthesis
- PMID: 10430882
- PMCID: PMC17719
- DOI: 10.1073/pnas.96.16.8985
An aminoacyl-tRNA synthetase paralog with a catalytic role in histidine biosynthesis
Abstract
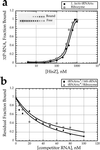
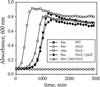
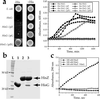
In addition to their essential catalytic role in protein biosynthesis, aminoacyl-tRNA synthetases participate in numerous other functions, including regulation of gene expression and amino acid biosynthesis via transamidation pathways. Herein, we describe a class of aminoacyl-tRNA synthetase-like (HisZ) proteins based on the catalytic core of the contemporary class II histidyl-tRNA synthetase whose members lack aminoacylation activity but are instead essential components of the first enzyme in histidine biosynthesis ATP phosphoribosyltransferase (HisG). Prediction of the function of HisZ in Lactococcus lactis was assisted by comparative genomics, a technique that revealed a link between the presence or the absence of HisZ and a systematic variation in the length of the HisG polypeptide. HisZ is required for histidine prototrophy, and three other lines of evidence support the direct involvement of HisZ in the transferase function. (i) Genetic experiments demonstrate that complementation of an in-frame deletion of HisG from Escherichia coli (which does not possess HisZ) requires both HisG and HisZ from L. lactis. (ii) Coelution of HisG and HisZ during affinity chromatography provides evidence of direct physical interaction. (iii) Both HisG and HisZ are required for catalysis of the ATP phosphoribosyltransferase reaction. This observation of a common protein domain linking amino acid biosynthesis and protein synthesis implies an early connection between the biosynthesis of amino acids and proteins.
Figures

References
-
- Bult C J, White D, Olsen G, Zhou L, Fleischmann R, Sutton G, Blake J, Fitzgerald L, Clayton R, Gocayne J, et al. Science. 1996;273:1058–1073. - PubMed
-
- Kunst F, Ogasawara N, Moszer I, Albertini A, Alloni G, Azeveda V, Bertero M, Bessieres P, Bolotin A, Borchert S, et al. Nature (London) 1997;390:249–256. - PubMed
-
- Deckert G, Warren P, Gaasterland T, Young W, Lenox A, Graham D, Overbeek R, Snead M, Keller M J, Aujay M, et al. Nature (London) 1998;392:353–358. - PubMed
-
- Curnow A W, Ibba M, Söll D. Nature (London) 1996;382:589–590. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

