A read-ahead function in archaeal DNA polymerases detects promutagenic template-strand uracil
- PMID: 10430892
- PMCID: PMC17729
- DOI: 10.1073/pnas.96.16.9045
A read-ahead function in archaeal DNA polymerases detects promutagenic template-strand uracil
Abstract
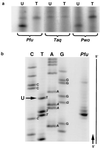
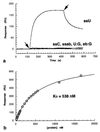
Deamination of cytosine to uracil is the most common promutagenic change in DNA, and it is greatly increased at the elevated growth temperatures of hyperthermophilic archaea. If not repaired to cytosine prior to replication, uracil in a template strand directs incorporation of adenine, generating a G.C --> A.U transition mutation in half the progeny. Surprisingly, genomic analysis of archaea has so far failed to reveal any homologues of either of the known families of uracil-DNA glycosylases responsible for initiating the base-excision repair of uracil in DNA, which is otherwise universal. Here we show that DNA polymerases from several hyperthermophilic archaea (including Vent and Pfu) specifically recognize the presence of uracil in a template strand and stall DNA synthesis before mutagenic misincorporation of adenine. A specific template-checking function in a DNA polymerase has not been observed previously, and it may represent the first step in a pathway for the repair of cytosine deamination in archaea.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

