Understanding beta-hairpin formation
- PMID: 10430896
- PMCID: PMC17733
- DOI: 10.1073/pnas.96.16.9068
Understanding beta-hairpin formation
Abstract
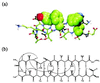
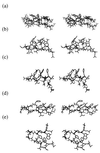
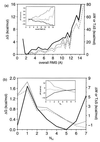
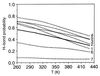
The kinetics of formation of protein structural motifs (e.g., alpha-helices and beta-hairpins) can provide information about the early events in protein folding. A recent study has used fluorescence measurements to monitor the folding thermodynamics and kinetics of a 16-residue beta-hairpin. In the present paper, we obtain the free energy surface and conformations involved in the folding of an atomistic model for the beta-hairpin from multicanonical Monte Carlo simulations. The results suggest that folding proceeds by a collapse that is downhill in free energy, followed by rearrangement to form a structure with part of the hydrophobic cluster; the hairpin hydrogen bonds propagate outwards in both directions from the partial cluster. Such a folding mechanism differs from the published interpretation of the experimental results, which is based on a helix-coil-type phenomenological model.
Figures

References
-
- Karplus M. Folding Des. 1997;2:S69–S75. - PubMed
-
- Dill K A, Chan H S. Nat Struct Biol. 1997;4:10–19. - PubMed
-
- Wolynes P G, Onuchic J N, Thirumalai D. Science. 1995;267:1619–1620. - PubMed
-
- Dobson C M, Karplus M, Šali A. Angew Chem Int Ed. 1998;37:868–893. - PubMed
-
- Dobson C M, Evans P A, Radford S E. Trends Biochem Sci. 1994;19:31–37. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

