Ligand-induced conformational changes observed in single RNA molecules
- PMID: 10430898
- PMCID: PMC17735
- DOI: 10.1073/pnas.96.16.9077
Ligand-induced conformational changes observed in single RNA molecules
Abstract
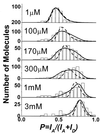
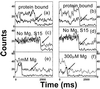
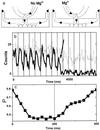
We present the first demonstration that fluorescence resonance energy transfer can be used to track the motion of a single molecule undergoing conformational changes. As a model system, the conformational changes of individual three-helix junction RNA molecules induced by the binding of ribosomal protein S15 or Mg(2+) ions were studied by changes in single-molecule fluorescence. The transition from an open to a folded configuration was monitored by the change of fluorescence resonance energy transfer between two different dye molecules attached to the ends of two helices in the RNA junction. Averaged behavior of RNA molecules closely resembles that of unlabeled molecules in solution determined by other bulk assays, proving that this approach is viable and suggesting new opportunities for studying protein-nucleic acids interactions. Surprisingly, we observed an anomalously broad distribution of RNA conformations at intermediate ion concentrations that may be attributed to foldability differences among RNA molecules. In addition, an experimental scheme was developed where the real-time response of single molecules can be followed under changing environments. As a demonstration, we repeatedly changed Mg(2+) concentration in the buffer while monitoring single RNA molecules and showed that individual RNA molecules can measure the instantaneous Mg(2+) concentration with 20-ms time resolution, making it the world's smallest Mg(2+) meter.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

