Digital PCR
- PMID: 10430926
- PMCID: PMC17763
- DOI: 10.1073/pnas.96.16.9236
Digital PCR
Abstract
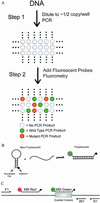
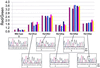
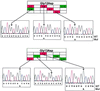
The identification of predefined mutations expected to be present in a minor fraction of a cell population is important for a variety of basic research and clinical applications. Here, we describe an approach for transforming the exponential, analog nature of the PCR into a linear, digital signal suitable for this purpose. Single molecules are isolated by dilution and individually amplified by PCR; each product is then analyzed separately for the presence of mutations by using fluorescent probes. The feasibility of the approach is demonstrated through the detection of a mutant ras oncogene in the stool of patients with colorectal cancer. The process provides a reliable and quantitative measure of the proportion of variant sequences within a DNA sample.
Figures


References
-
- Vogelstein B V, Kinzler K W. The Genetic Basis of Human Cancer. Toronto: McGraw–Hill; 1998.
-
- Lee C M, Weindruch R, Aiken J M. Free Radical Biol Med. 1997;22:1259–1269. - PubMed
-
- Ozawa T. Physiol Rev. 1997;77:425–464. - PubMed
-
- Sidransky D, Von Eschenbach A, Tsai Y C, Jones P, Summerhayes I, Marshall F, Paul M, Green P, Hamilton S R, Frost P, et al. Science. 1991;252:706–709. - PubMed
-
- Sidransky D, Tokino T, Hamilton S R, Kinzler K W, Levin B, Frost P, Vogelstein B. Science. 1992;256:102–105. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

