Fine-mapping of quantitative trait loci by identity by descent in outbred populations: application to milk production in dairy cattle
- PMID: 10430929
- PMCID: PMC17766
- DOI: 10.1073/pnas.96.16.9252
Fine-mapping of quantitative trait loci by identity by descent in outbred populations: application to milk production in dairy cattle
Abstract
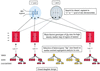
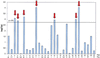
We previously mapped a quantitative trait locus (QTL) affecting milk production to bovine chromosome 14. To refine the map position of this QTL, we have increased the density of the genetic map of BTA14q11-16 by addition of nine microsatellites and three single nucleotide polymorphisms. Fine-mapping of the QTL was accomplished by a two-tiered approach. In the first phase, we identified seven sires heterozygous "Qq" for the QTL by marker-assisted segregation analysis in a Holstein-Friesian pedigree comprising 1,158 individuals. In a second phase, we genotyped the seven selected sires for the newly developed high-density marker map and searched for a shared haplotype flanking an hypothetical, identical-by-descent QTL allele with large substitution effect. The seven chromosomes increasing milk fat percentage were indeed shown to carry a common chromosome segment with an estimated size of 5 cM predicted to contain the studied QTL. The same haplotype was shown to be associated with increased fat percentage in the general population as well, providing additional support in favor of the location of the QTL within the corresponding interval.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

