A single gene (tts) located outside the cap locus directs the formation of Streptococcus pneumoniae type 37 capsular polysaccharide. Type 37 pneumococci are natural, genetically binary strains
- PMID: 10432287
- PMCID: PMC2195575
- DOI: 10.1084/jem.190.2.241
A single gene (tts) located outside the cap locus directs the formation of Streptococcus pneumoniae type 37 capsular polysaccharide. Type 37 pneumococci are natural, genetically binary strains
Abstract
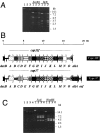
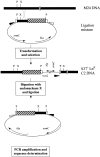
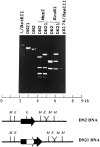
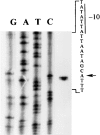
The molecular aspects of the type 37 pneumococcal capsular biosynthesis, a homopolysaccharide composed of sophorosyl units (beta-d-Glc-(1-->2)-beta-d-Glc) linked by beta-1,3 bonds, have been studied. Remarkably, the biosynthesis of the type 37 capsule is driven by a single gene (tts) located far apart from the cap locus responsible for capsular formation in all of the types characterized to date in Streptococcus pneumoniae. However, a cap37 locus virtually identical to the cap33f cluster has been found in type 37 strains, although some of its genes are inactivated by mutations. The tts gene has been sequenced and its transcription start point determined. Tts shows sequence motifs characteristic of cellulose synthases and other beta-glycosyltransferases. Insertion of the tts gene into the pneumococcal DNA causes a noticeable genome reorganization in such a way that genes normally separated by more than 350 kb in the chromosome are located together in clinical isolates of type 37. Encapsulated pneumococcal strains belonging to 10 different serotypes (or serogroups) transformed with tts synthesized type 37 polysaccharide, leading to the formation of strains that display the binary type of capsule. Type 37 pneumococcus constitutes the first case of a natural, genetically binary strain and represents a novel alternative to the mechanisms of intertype transformation.
Figures



Similar articles
-
Tts, a processive beta-glucosyltransferase of Streptococcus pneumoniae, directs the synthesis of the branched type 37 capsular polysaccharide in Pneumococcus and other gram-positive species.J Biol Chem. 2001 Jun 15;276(24):21053-61. doi: 10.1074/jbc.M010287200. Epub 2001 Mar 22. J Biol Chem. 2001. PMID: 11264282
-
A functional analysis of the Streptococcus pneumoniae genes involved in the synthesis of type 1 and type 3 capsular polysaccharides.Microb Drug Resist. 1997 Spring;3(1):73-88. doi: 10.1089/mdr.1997.3.73. Microb Drug Resist. 1997. PMID: 9109098 Review.
-
Molecular organization of the genes required for the synthesis of type 1 capsular polysaccharide of Streptococcus pneumoniae: formation of binary encapsulated pneumococci and identification of cryptic dTDP-rhamnose biosynthesis genes.Mol Microbiol. 1997 Jul;25(1):79-92. doi: 10.1046/j.1365-2958.1997.4341801.x. Mol Microbiol. 1997. PMID: 11902728
-
Current trends in capsular polysaccharide biosynthesis of Streptococcus pneumoniae.Res Microbiol. 2000 Jul-Aug;151(6):429-35. doi: 10.1016/s0923-2508(00)00173-x. Res Microbiol. 2000. PMID: 10961455 Review.
-
Characterization of the cassette containing genes for type 3 capsular polysaccharide biosynthesis in Streptococcus pneumoniae.J Exp Med. 1995 Mar 1;181(3):973-83. doi: 10.1084/jem.181.3.973. J Exp Med. 1995. PMID: 7869055 Free PMC article.
Cited by
-
Heterologous expression of a position 2-substituted (1-->3)-beta-D-glucan in Lactococcus lactis.Appl Environ Microbiol. 2008 Aug;74(16):5259-62. doi: 10.1128/AEM.00463-08. Epub 2008 Jun 20. Appl Environ Microbiol. 2008. PMID: 18567684 Free PMC article.
-
Influence of Different Sugars and Initial pH on β-Glucan Formation by Lactobacillus brevis TMW 1.2112.Curr Microbiol. 2018 Jul;75(7):794-802. doi: 10.1007/s00284-018-1450-z. Epub 2018 Feb 13. Curr Microbiol. 2018. PMID: 29442149
-
Population-based analysis of invasive nontypeable pneumococci reveals that most have defective capsule synthesis genes.PLoS One. 2014 May 15;9(5):e97825. doi: 10.1371/journal.pone.0097825. eCollection 2014. PLoS One. 2014. PMID: 24831650 Free PMC article.
-
Biofilm formation by Streptococcus pneumoniae: role of choline, extracellular DNA, and capsular polysaccharide in microbial accretion.J Bacteriol. 2006 Nov;188(22):7785-95. doi: 10.1128/JB.00673-06. Epub 2006 Aug 25. J Bacteriol. 2006. PMID: 16936041 Free PMC article.
-
SP_0916 Is an Arginine Decarboxylase That Catalyzes the Synthesis of Agmatine, Which Is Critical for Capsule Biosynthesis in Streptococcus pneumoniae.Front Microbiol. 2020 Sep 18;11:578533. doi: 10.3389/fmicb.2020.578533. eCollection 2020. Front Microbiol. 2020. PMID: 33072045 Free PMC article.
References
-
- Galán J.E., Bliska J.B. Cross-talk between bacterial pathogens and their host cells. Annu. Rev. Cell Dev. Biol. 1996;12:221–255. - PubMed
-
- Feldman C., Klugman K.P. Pneumococcal infections. Curr. Opin. Infect. Dis. 1997;10:109–115.
-
- Gardner P., Schaffner W. Immunization of adults. N. Engl. J. Med. 1993;328:1252–1258. - PubMed
-
- Muñoz R., Mollerach M., López R., García E. Molecular organization of the genes required for the synthesis of type 1 capsular polysaccharide of Streptococcus pneumoniaeformation of binary encapsulated pneumococci and identification of cryptic dTDP-rhamnose biosynthesis genes. Mol. Microbiol. 1997;25:79–92. - PubMed
-
- Arrecubieta C., García E., López R. Sequence and transcriptional analysis of a DNA region involved in the production of capsular polysaccharide in Streptococcus pneumoniae type 3. Gene. 1995;167:1–7. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

