Protein kinase activity and identification of a toxic effector domain of the target of rapamycin TOR proteins in yeast
- PMID: 10436010
- PMCID: PMC25485
- DOI: 10.1091/mbc.10.8.2531
Protein kinase activity and identification of a toxic effector domain of the target of rapamycin TOR proteins in yeast
Abstract
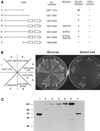
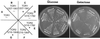
In complex with FKBP12, the immunosuppressant rapamycin binds to and inhibits the yeast TOR1 and TOR2 proteins and the mammalian homologue mTOR/FRAP/RAFT1. The TOR proteins promote cell cycle progression in yeast and human cells by regulating translation and polarization of the actin cytoskeleton. A C-terminal domain of the TOR proteins shares identity with protein and lipid kinases, but only one substrate (PHAS-I), and no regulators of the TOR-signaling cascade have been identified. We report here that yeast TOR1 has an intrinsic protein kinase activity capable of phosphorylating PHAS-1, and this activity is abolished by an active site mutation and inhibited by FKBP12-rapamycin or wortmannin. We find that an intact TOR1 kinase domain is essential for TOR1 functions in yeast. Overexpression of a TOR1 kinase-inactive mutant, or of a central region of the TOR proteins distinct from the FRB and kinase domains, was toxic in yeast, and overexpression of wild-type TOR1 suppressed this toxic effect. Expression of the TOR-toxic domain leads to a G1 cell cycle arrest, consistent with an inhibition of TOR function in translation. Overexpression of the PLC1 gene, which encodes the yeast phospholipase C homologue, suppressed growth inhibition by the TOR-toxic domains. In conclusion, our findings identify a toxic effector domain of the TOR proteins that may interact with substrates or regulators of the TOR kinase cascade and that shares sequence identity with other PIK family members, including ATR, Rad3, Mei-41, and ATM.
Figures



References
-
- Alarcon CM, Cardenas ME, Heitman J. Mammalian RAFT1 kinase domain provides rapamycin-sensitive TOR function in yeast. Genes Dev. 1996;10:279–288. - PubMed
-
- Barlow C, et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86:159–171. - PubMed
-
- Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–759. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

