The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins
- PMID: 10436016
- PMCID: PMC25492
- DOI: 10.1091/mbc.10.8.2607
The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins
Abstract
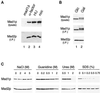
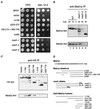
The spindle checkpoint arrests the cell cycle at metaphase in the presence of defects in the mitotic spindle or in the attachment of chromosomes to the spindle. When spindle assembly is disrupted, the budding yeast mad and bub mutants fail to arrest and rapidly lose viability. We have cloned the MAD2 gene, which encodes a protein of 196 amino acids that remains at a constant level during the cell cycle. Gel filtration and co-immunoprecipitation analyses reveal that Mad2p tightly associates with another spindle checkpoint component, Mad1p. This association is independent of cell cycle stage and the presence or absence of other known checkpoint proteins. In addition, Mad2p binds to all of the different phosphorylated isoforms of Mad1p that can be resolved on SDS-PAGE. Deletion and mutational analysis of both proteins indicate that association of Mad2p with Mad1p is critical for checkpoint function and for hyperphosphorylation of Mad1p.
Figures






References
-
- Aravind L, Koonin EV. The HORMA domain: a common structural denominator in mitotic checkpoints, chromosome synapsis and DNA repair. Trends Biochem Sci. 1998;23:284–286. - PubMed
-
- Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. - PubMed
-
- Chen R-H, Juo P-C, Curran T, Blenis J. Phosphorylation of c-Fos at the C-terminus enhances its transforming activity. Oncogene. 1996;12:1493–1502. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

