Excision of IS492 requires flanking target sequences and results in circle formation in Pseudoalteromonas atlantica
- PMID: 10438765
- PMCID: PMC93982
- DOI: 10.1128/JB.181.16.4937-4948.1999
Excision of IS492 requires flanking target sequences and results in circle formation in Pseudoalteromonas atlantica
Abstract
The gram-negative marine bacterium Pseudoalteromonas atlantica produces extracellular polysaccharide (EPS) that is important in biofilm formation by this bacterium. Insertion and precise excision of IS492 at a locus essential for extracellular polysaccharide production (eps) controls phase variation of EPS production in P. atlantica. Examination of IS492 transposition in P. atlantica by using a PCR-based assay revealed a circular form of IS492 that may be an intermediate in transposition or a terminal product of excision. The DNA sequence of the IS492 circle junction indicates that the ends of the element are juxtaposed with a 5-bp spacer sequence. This spacer sequence corresponds to the 5-bp duplication of the chromosomal target sequence found at all IS492 insertion sites on the P. atlantica chromosome that we identified by using inverse PCR. IS492 circle formation correlated with precise excision of IS492 from the P. atlantica eps target sequence when introduced into Escherichia coli on a plasmid. Deletion analyses of the flanking host sequences at the eps insertion site for IS492 demonstrated that the 5-bp duplicated target sequence is essential for precise excision of IS492 and circle formation in E. coli. Excision of IS492 in E. coli also depends on the level of expression of the putative transposase, MooV. A regulatory role for the circular form of IS492 is suggested by the creation of a new strong promoter for expression of mooV by the joining of the ends of the insertion sequence element at the circle junction.
Figures






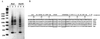

References
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley; 1993.
-
- Chan K Y, Baumann L, Garza M M, Baumann P. Two new species of Alteromonas: Alteromonas espejiana and Alteromonas undina. Int J Syst Bacteriol. 1978;28:217–222.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

