The bvr locus of Listeria monocytogenes mediates virulence gene repression by beta-glucosides
- PMID: 10438775
- PMCID: PMC93992
- DOI: 10.1128/JB.181.16.5024-5032.1999
The bvr locus of Listeria monocytogenes mediates virulence gene repression by beta-glucosides
Abstract
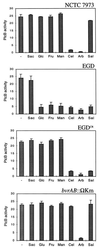
The beta-glucoside cellobiose has been reported to specifically repress the PrfA-dependent virulence genes hly and plcA in Listeria monocytogenes NCTC 7973. This led to the hypothesis that beta-glucosides, sugars of plant origin, may act as signal molecules, preventing the expression of virulence genes if L. monocytogenes is living in its natural habitat (soil). In three other laboratory strains (EGD, L028, and 10403S), however, the effect of cellobiose was not unique, and all fermentable carbohydrates repressed hly. This suggested that the downregulation of virulence genes by beta-glucosides is not a specific phenomenon but, rather, an aspect of a global regulatory mechanism of catabolite repression (CR). We assessed the effect of carbohydrates on virulence gene expression in a panel of wild-type isolates of L. monocytogenes by using the PrfA-dependent phospholipase C gene plcB as a reporter. Utilization of any fermentable sugar caused plcB repression in wild-type L. monocytogenes. However, an EGD variant was identified in which, as in NCTC 7973, plcB was only repressed by beta-glucosides. Thus, the regulation of L. monocytogenes virulence genes by sugars appears to be mediated by two separate mechanisms, one presumably involving a CR pathway and another specifically responding to beta-glucosides. We have identified in L. monocytogenes a 4-kb operon, bvrABC, encoding an antiterminator of the BglG family (bvrA), a beta-glucoside-specific enzyme II permease component of the phosphoenolpyruvate-sugar phosphotransferase system (bvrB), and a putative ADP-ribosylglycohydrolase (bvrC). Low-stringency Southern blots showed that this locus is absent from other Listeria spp. Transcription of bvrB was induced by cellobiose and salicin but not by arbutin. Disruption of the bvr operon by replacing part of bvrAB with an interposon abolished the repression by cellobiose and salicin but not that by arbutin. Our data indicate that the bvr locus encodes a beta-glucoside-specific sensor that mediates virulence gene repression upon detection of cellobiose and salicin. Bvr is the first sensory system found in L. monocytogenes that is involved in environmental regulation of virulence genes.
Figures




References
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1991.
-
- Bohne J, Sokolovic Z, Goebel W. Transcriptional regulation of prfA and PrfA-regulated virulence genes in Listeria monocytogenes. Mol Microbiol. 1994;11:1141–1150. - PubMed
-
- Brehm K, Haas A, Goebel W, Kreft J. A gene encoding a superoxide dismutase of the facultative intracellular bacterium Listeria monocytogenes. Gene. 1992;118:121–125. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

