The herpes simplex virus vhs protein induces endoribonucleolytic cleavage of target RNAs in cell extracts
- PMID: 10438802
- PMCID: PMC104239
- DOI: 10.1128/JVI.73.9.7153-7164.1999
The herpes simplex virus vhs protein induces endoribonucleolytic cleavage of target RNAs in cell extracts
Abstract
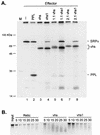
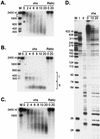
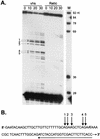
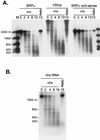
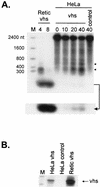
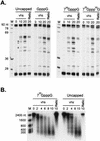
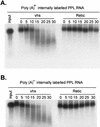
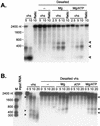
The herpes simplex virus virion host shutoff (vhs) protein (UL41 gene product) is a component of the HSV virion tegument that triggers shutoff of host protein synthesis and accelerated mRNA degradation during the early stages of HSV infection. Previous studies have demonstrated that extracts from HSV-infected cells and partially purified HSV virions display vhs-dependent RNase activity and that vhs is sufficient to trigger accelerated RNA degradation when expressed as the only HSV protein in an in vitro translation system derived from rabbit reticulocytes. We have used the rabbit reticulocyte translation system to characterize the mode of vhs-induced RNA decay in more detail. We report here that vhs-dependent RNA decay proceeds through endoribonucleolytic cleavage, is not affected by the presence of a 5' cap or a 3' poly(A) tail in the RNA substrate, requires Mg(2+), and occurs in the absence of ribosomes. Intriguingly, sites of preferential initial cleavage were clustered over the 5' quadrant of one RNA substrate that was characterized in detail. The vhs homologue of pseudorabies virus also induced accelerated RNA decay in this in vitro system.
Figures

References
-
- Beelman C A, Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. - PubMed
-
- Ben-Porat T, Rakusanova T, Kaplan A S. Early functions of the genome of herpesvirus. II. Inhibition of formation of cell-specific polysomes. Virology. 1971;46:890–899. - PubMed
-
- Berthomme H, Jacquemont B, Epstein A. The pseudorabies virus host-shutoff homolog gene: nucleotide sequence and comparison with alphaherpesvirus protein counterparts. Virology. 1993;193:1028–1032. - PubMed
-
- Brown B D, Zipkin I D, Harland R M. Sequence-specific endonucleolytic cleavage and protection of mRNA in Xenopus and Drosophila. Genes Dev. 1993;7:1620–1631. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

