An inducible human immunodeficiency virus type 1 (HIV-1) vector which effectively suppresses HIV-1 replication
- PMID: 10438857
- PMCID: PMC104294
- DOI: 10.1128/JVI.73.9.7671-7677.1999
An inducible human immunodeficiency virus type 1 (HIV-1) vector which effectively suppresses HIV-1 replication
Abstract
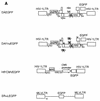
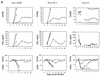
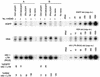
Recently, gene therapy vectors based upon the human immunodeficiency virus type 1 (HIV-1) genome have been developed. Here, we create an HIV-1 vector which is defective for all HIV-1 genes, but which maintains cis-acting elements required for efficient packaging, infection, and expression. In T cells transduced by this vector, vector expression is low but efficiently induced following HIV-1 infection. Remarkably, although the HIV-1 vector does not contain specific anti-HIV-1 therapeutic genes, the presence of the vector alone is sufficient to inhibit the spread of HIV-1 infection. The mechanism of inhibition is likely to be at the level of competition for limiting substrates required for either efficient packaging or reverse transcription, thereby selecting against propagation of wild-type HIV-1. These results provide proof of a concept for potential application of a novel HIV-1 vector in HIV-1 disease.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

