Evolution and horizontal transfer of dUTPase-encoding genes in viruses and their hosts
- PMID: 10438861
- PMCID: PMC104298
- DOI: 10.1128/JVI.73.9.7710-7721.1999
Evolution and horizontal transfer of dUTPase-encoding genes in viruses and their hosts
Abstract
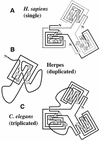
dUTPase is a ubiquitous and essential enzyme responsible for regulating cellular levels of dUTP. The dut gene exists as single, tandemly duplicated, and tandemly triplicated copies. Crystallized single-copy dUTPases have been shown to assemble as homotrimers. dUTPase is encoded as an auxiliary gene in a number of virus genomes. The origin of viral dut genes has remained unresolved since their initial discovery. A comprehensive analysis of dUTPase amino acid sequence relationships was performed to explore the evolutionary dynamics of dut in viruses and their hosts. Our data set, comprised of 24 host and 51 viral sequences, includes representative sequences from available eukaryotes, archaea, eubacteria cells, and viruses, including herpesviruses. These amino acid sequences were aligned by using a hidden Markov model approach developed to align divergent data. Known secondary structures from single-copy crystals were mapped onto the aligned duplicate and triplicate sequences. We show how duplicated dUTPases might fold into a monomer, and we hypothesize that triplicated dUTPases also assemble as monomers. Phylogenetic analysis revealed at least five viral dUTPase sequence lineages in well-supported monophyletic clusters with eukaryotic, eubacterial, and archaeal hosts. We have identified all five as strong examples of horizontal transfer as well as additional potential transfer of dut genes among eubacteria, between eubacteria and viruses, and between retroviruses. The evidence for horizontal transfers is particularly interesting since eukaryotic dut genes have introns, while DNA virus dut genes do not. This implies that an intermediary retroid agent facilitated the horizontal transfer process between host mRNA and DNA viruses.
Figures

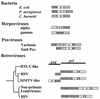
 , the DNA synthesis flavoprotein;
, the DNA synthesis flavoprotein;  , unknown function;
, unknown function;  , the phosphomannomutase;
, the phosphomannomutase;  , the outer membrane protein PIB. Herpesvirus genes:
, the outer membrane protein PIB. Herpesvirus genes:  , the ribonucleotide reductase-related protein;
, the ribonucleotide reductase-related protein;  , the primase. Poxviruses:
, the primase. Poxviruses:  , the transcription initiation factor;
, the transcription initiation factor;  , the ribonucleotide reductase. Retroviruses:
, the ribonucleotide reductase. Retroviruses:  , the protease;
, the protease;  , the reverse transcriptase;
, the reverse transcriptase;  , the ribonuclease H;
, the ribonuclease H;  , the integrase. Unidentified homologous reading frames are indicated (
, the integrase. Unidentified homologous reading frames are indicated ( ). Unidentified nonhomologous reading frames are indicated with a question mark.
). Unidentified nonhomologous reading frames are indicated with a question mark.

References
-
- Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Baer R, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatfull G, Hudson G S, Satchwell S C, Seguin C. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. - PubMed
-
- Caradonna S J, Adamkiewicz D M. Purification and properties of the deoxyuridine triphosphate nucleotidohydrolase enzyme derived from HeLa S3 cells. J Biol Chem. 1984;259:5459–5464. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

