Uncoupling of the hnRNP Npl3p from mRNAs during the stress-induced block in mRNA export
- PMID: 10444597
- PMCID: PMC316916
- DOI: 10.1101/gad.13.15.1994
Uncoupling of the hnRNP Npl3p from mRNAs during the stress-induced block in mRNA export
Abstract
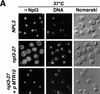
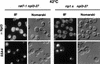
Npl3p, the major mRNA-binding protein of the yeast Saccharomyces cerevisiae shuttles between the nucleus and the cytoplasm. A single amino acid change in the carboxyl terminus of Npl3p (E409 --> K) renders the mutant protein largely cytoplasmic because of a delay in its import into the nucleus. This import defect can be reversed by increasing the intracellular concentration of Mtr10p, the nuclear import receptor for Npl3p. Conversely, using this mutant, we show that Npl3p and mRNA export out of the nucleus is significantly slowed in cells bearing mutations in XPO1/CRM1, which encodes the export receptor for NES-containing proteins and in RAT7, which encodes an essential nucleoporin. Interestingly, following induction of stress by heat shock, high salt, or ethanol, conditions under which most mRNA export is blocked, Npl3p is still exported from the nucleus. The stress-induced export of Npl3p is independent of both the activity of Xpo1p and the continued selective export of heat-shock mRNAs that occurs following stress. UV-cross-linking experiments show that Npl3p is bound to mRNA under normal conditions, but is no longer RNA associated in stressed cells. Taken together, we suggest that the uncoupling of Npl3p and possibly other mRNA-binding proteins from mRNAs in the nucleus provides a general switch that regulates mRNA export. By this model, under normal conditions Npl3p is a major component of an export-competent RNP complex. However, under conditions of stress, Npl3p no longer associates with the export complex, rendering it export incompetent and thus nuclear.
Figures










References
-
- Amberg DC, Goldstein AL, Cole CN. Isolation and characterization of RAT1: An essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes & Dev. 1992;6:1173–1189. - PubMed
-
- Bogerd HP, Echarri A, Ross TM, Cullen BR. Inhibition of human immunodeficiency virus rev and human T-cell leukemia virus rex function, but not mason-pfizer monkey virus constitutive transport element activity, by a mutant human nucleoporin targeted to crm1. J Virol. 1998;72:8627–8635. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
