High-resolution landmark framework for the sequence-ready mapping of Xq23-q26.1
- PMID: 10447510
- PMCID: PMC310799
High-resolution landmark framework for the sequence-ready mapping of Xq23-q26.1
Abstract
We have established a landmark framework map over 20-25 Mb of the long arm of the human X chromosome using yeast artificial chromosome (YAC) clones. The map has approximately one landmark per 45 kb of DNA and stretches from DXS7531 in proximal Xq23 to DXS895 in proximal Xq26, connecting to published framework maps on its proximal and distal sides. There are three gaps in the framework map resulting from the failure to obtain clone coverage from the YAC resources available. Estimates of the maximum sizes of these gaps have been obtained. The four YAC contigs have been positioned and oriented using somatic-cell hybrids and fluorescence in situ hybridization, and the largest is estimated to cover approximately 15 Mb of DNA. The framework map is being used to assemble a sequence-ready map in large-insert bacterial clones, as part of an international effort to complete the sequence of the X chromosome. PAC and BAC contigs currently cover 18 Mb of the region, and from these, 12 Mb of finished sequence is available.
Figures





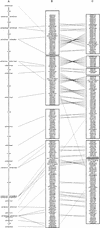

References
-
- Bentley DR, Todd C, Collins J, Holland J, Dunham I, Hassock S, Bankier A, Gianelli F. The development and application of automated gridding for efficient screening of yeast and bacterial ordered libraries. Genomics. 1992;12:534–541. - PubMed
-
- Bentley DR, Pruitt KD, Deloukas P, Schuler GD, Ostell J. Coordination of human genome sequencing via a consensus framework map. Trends Genet. 1998;14:381–384. - PubMed
-
- Brownstein BH, Silverman GA, Little RD, Burke DT, Korsmeyer SJ, Schlessinger D, Olson MV. Isolation of single-copy human genes from a library of yeast artificial chromosome clones. Science. 1989;244:1348–1351. - PubMed
-
- Buetow KH, Weber JL, Ludwigsen S, Scherpbier-Heddema T, Duyk GM, Sheffield VC, Wang Z, Murray JC. Integrated human genome-wide maps constructed using the CEPH reference panel. Nat Genet. 1994;6:391–393. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
