Genotypes of canine distemper virus determined by analysis of the hemagglutinin genes of recent isolates from dogs in Japan
- PMID: 10449479
- PMCID: PMC85418
- DOI: 10.1128/JCM.37.9.2936-2942.1999
Genotypes of canine distemper virus determined by analysis of the hemagglutinin genes of recent isolates from dogs in Japan
Abstract
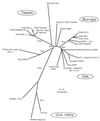
Canine distemper of domestic dogs is caused by canine distemper virus (CDV), a member of the morbilliviruses. It has been a highly contagious disease of great veterinary importance for centuries, but for the last several decades it has been controlled satisfactorily by modified live vaccines. In the 1990s, however, it was described that CDV strains genetically different from vaccine strains may have caused the disease in vaccinated dogs. The highest antigenic variation is found in the H protein. Therefore, in the present study, hemagglutinin (H) genes obtained from current vaccines and field isolates and amplified directly from clinical specimens were genetically analyzed by restriction fragment length polymorphism assay and sequencing. Phylogenetic analysis of the H-gene amino acid sequences revealed that at least two CDV genotypes are circulating among dogs in Japan; one is a genotype to which almost all Japanese CDV isolates belong and the other has not been previously described. Both are separate and independent from the other lineages or genotypes of vaccine strains, as well as European and U.S. CDV isolates. The results suggest that CDV has also evolved in Japan, and further studies will be needed for an evaluation and possible improvement of the efficacies of current CDV vaccines.
Figures


References
-
- Appel M. Canine distemper virus. In: Appel M J, editor. Virus infections of carnivores. Amsterdam, The Netherlands: Elsevier Science Publishers B.V.; 1987. pp. 133–159.
-
- Appel M J G, Summers B A. Pathogenicity of morbilliviruses for terrestrial carnivores. Vet Microbiol. 1995;44:187–191. - PubMed
-
- Appel M J G, Yates R A, Foley G L, Bernstein J J, Santinelli S, Spelman L H, Miller L D, Arp L H, Anderson M, Barr M, Pearce-Kelling S, Summers B A. Canine distemper epizootic in lions, tigers, and leopards in North America. J Vet Diagn Invest. 1994;6:277–288. - PubMed
-
- Barrett T, Clarke D K, Evans S A, Rima B K. The nucleotide sequence of the gene encoding the F protein of canine distemper virus: a comparison of the deduced amino acid sequences with other paramyxoviruses. Virus Res. 1987;8:373–386. - PubMed
-
- Blixenkrone-Møller M, Svansson V, Appel M, Krogsrud J, Have P, Orvell C. Antigenic relationship between field isolates of morbilliviruses from different carnivores. Arch Virol. 1992;123:279–294. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

