Protein design is a key factor for subunit-subunit association
- PMID: 10449742
- PMCID: PMC22258
- DOI: 10.1073/pnas.96.17.9616
Protein design is a key factor for subunit-subunit association
Abstract
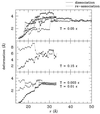
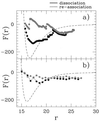
Fundamental questions about role of the quaternary structures are addressed by using a statistical mechanics off-lattice model of a dimer protein. The model, in spite of its simplicity, captures key features of the monomer-monomer interactions revealed by atomic force experiments. Force curves during association and dissociation processes are characterized by sudden jumps followed by smooth behavior and form hysteresis loops. Furthermore, the process is reversible in a finite range of temperature stabilizing the dimer, and the width of the hysteresis loop increases as the design procedure improves: i.e., stabilizes the dimer more. It is shown that, in the interface between the two monomeric subunits, the design procedure naturally favors those amino acids whose mutual interaction is stronger.
Figures





References
-
- Schultz G E, Schirmer R H. Principles of Protein Structure. New York: Springer; 1979.
-
- Whittingham J L, Edwards D J, Antson A A, Clarkson J M, Dodson G G. Biochemistry. 1998;37:11516–11523. - PubMed
-
- Wlodawer A, Vondrasek J. Annu Rev Biophys Biomol Struct. 1998;27:249–284. - PubMed
-
- McKenzie H A. Adv Protein Chem. 1967;22:55–234. - PubMed
-
- Pocker Y, Biswas S B. Biochemistry. 1981;20:4354–4361. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

