Basonuclin, a zinc finger protein of keratinocytes and reproductive germ cells, binds to the rRNA gene promoter
- PMID: 10449744
- PMCID: PMC22260
- DOI: 10.1073/pnas.96.17.9628
Basonuclin, a zinc finger protein of keratinocytes and reproductive germ cells, binds to the rRNA gene promoter
Abstract
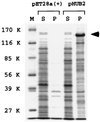
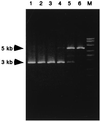
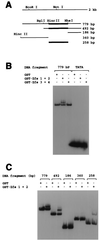
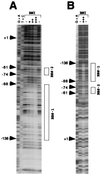
Basonuclin is a protein containing three pairs of C(2)H(2) zinc fingers. The protein has been found in the basal (germinal) cell layer of stratified squamous epithelia, such as the epidermis, and in germ cells of the testis and ovary. We show here that the human protein has specific affinity for a segment of the promoter of the gene for rRNA. Basonuclin interacts with two separate parts of the promoter, each possessing dyad symmetry. The upstream part, but not the downstream part, is known to bind UBF1, a transcription factor for rDNA. Basonuclin is likely to be a cell-type-specific regulatory protein for rDNA transcription.
Figures
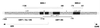
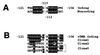
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

