Mapping the molecular determinant of pathogenicity in a hammerhead viroid: a tetraloop within the in vivo branched RNA conformation
- PMID: 10449802
- PMCID: PMC22318
- DOI: 10.1073/pnas.96.17.9960
Mapping the molecular determinant of pathogenicity in a hammerhead viroid: a tetraloop within the in vivo branched RNA conformation
Abstract
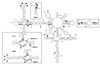
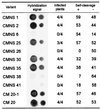
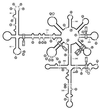
Chrysanthemum chlorotic mottle viroid (CChMVd) is an RNA of 398-399 nt that can adopt hammerhead structures in both polarity strands. We have identified by Northern-blot hybridization a nonsymptomatic strain (CChMVd-NS) that protects against challenge inoculation with the symptomatic strain (CChMVd-S). Analysis of CChMVd-NS cDNA clones has revealed a size and sequence very similar to those of the CChMVd-S strain. Some of the mutations observed in CChMVd-NS molecular variants were previously identified in CChMVd-S RNA, but others were never found in this RNA. When bioassayed in chrysanthemum, cDNA clones containing the CChMVd-NS specific mutations were infectious but nonsymptomatic. Site-directed mutagenesis showed that one of the CChMVd-NS-specific mutations, a UUUC --> GAAA substitution, was sufficient to change the symptomatic phenotype into the nonsymptomatic one without altering the final accumulation level of the viroid RNA. The pathogenicity determinant-to our knowledge, a determinant of this class has not been described previously in hammerhead viroids-is located in a tetraloop of the computer-predicted branched conformation for CChMVd RNA. Analysis of the sequence heterogeneity found in CChMVd-S and -NS variants strongly supports the existence of such a conformation in vivo, showing that the rod-like or quasi-rod-like secondary structure is not a universal paradigm for viroids.
Figures


Similar articles
-
Chrysanthemum chlorotic mottle viroid: unusual structural properties of a subgroup of self-cleaving viroids with hammerhead ribozymes.Proc Natl Acad Sci U S A. 1997 Oct 14;94(21):11262-7. doi: 10.1073/pnas.94.21.11262. Proc Natl Acad Sci U S A. 1997. PMID: 9326597 Free PMC article.
-
A kissing-loop interaction in a hammerhead viroid RNA critical for its in vitro folding and in vivo viability.RNA. 2005 Jul;11(7):1073-83. doi: 10.1261/rna.2230605. Epub 2005 May 31. RNA. 2005. PMID: 15928342 Free PMC article.
-
Chrysanthemum chlorotic mottle viroid RNA: dissection of the pathogenicity determinant and comparative fitness of symptomatic and non-symptomatic variants.J Mol Biol. 2002 Aug 16;321(3):411-21. doi: 10.1016/s0022-2836(02)00629-0. J Mol Biol. 2002. PMID: 12162955
-
Viroids with hammerhead ribozymes: some unique structural and functional aspects with respect to other members of the group.Biol Chem. 1999 Jul-Aug;380(7-8):849-54. doi: 10.1515/BC.1999.104. Biol Chem. 1999. PMID: 10494833 Review.
-
A current overview of two viroids that infect chrysanthemums: Chrysanthemum stunt viroid and Chrysanthemum chlorotic mottle viroid.Viruses. 2013 Apr 17;5(4):1099-113. doi: 10.3390/v5041099. Viruses. 2013. PMID: 23594461 Free PMC article. Review.
Cited by
-
Viroid pathogenicity: one process, many faces.Viruses. 2009 Sep;1(2):298-316. doi: 10.3390/v1020298. Epub 2009 Sep 10. Viruses. 2009. PMID: 21994551 Free PMC article.
-
Molecular diversity among viroids infecting chrysanthemum in India.Virus Genes. 2017 Aug;53(4):636-642. doi: 10.1007/s11262-017-1468-5. Epub 2017 May 19. Virus Genes. 2017. PMID: 28527099
-
The transcription initiation sites of eggplant latent viroid strands map within distinct motifs in their in vivo RNA conformations.RNA Biol. 2016;13(1):83-97. doi: 10.1080/15476286.2015.1119365. RNA Biol. 2016. PMID: 26618399 Free PMC article.
-
Involvement of the chloroplastic isoform of tRNA ligase in the replication of viroids belonging to the family Avsunviroidae.J Virol. 2012 Aug;86(15):8269-76. doi: 10.1128/JVI.00629-12. Epub 2012 May 23. J Virol. 2012. PMID: 22623792 Free PMC article.
-
Inhibition of cell growth and shoot development by a specific nucleotide sequence in a noncoding viroid RNA.Plant Cell. 2003 Jun;15(6):1360-74. doi: 10.1105/tpc.011585. Plant Cell. 2003. PMID: 12782729 Free PMC article.
References
-
- Diener T O. The Viroids. New York: Plenum; 1987.
-
- Flores R, Randles J W, Bar-Joseph M, Diener T O. Arch Virol. 1998;143:623–629. - PubMed
-
- Flores R, Di Serio F, Hernández C. Semin Virol. 1997;8:65–73.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

