The strength of interaction at the Raf cysteine-rich domain is a critical determinant of response of Raf to Ras family small GTPases
- PMID: 10454553
- PMCID: PMC84512
- DOI: 10.1128/MCB.19.9.6057
The strength of interaction at the Raf cysteine-rich domain is a critical determinant of response of Raf to Ras family small GTPases
Abstract
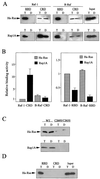
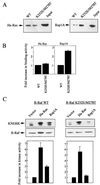
To be fully activated at the plasma membrane, Raf-1 must establish two distinct modes of interactions with Ras, one through its Ras-binding domain and the other through its cysteine-rich domain (CRD). The Ras homologue Rap1A is incapable of activating Raf-1 and even antagonizes Ras-dependent activation of Raf-1. We proposed previously that this property of Rap1A may be attributable to its greatly enhanced interaction with Raf-1 CRD compared to Ras. On the other hand, B-Raf, another Raf family member, is activatable by both Ras and Rap1A. When interactions with Ras and Rap1A were measured, B-Raf CRD did not exhibit the enhanced interaction with Rap1A, suggesting that the strength of interaction at CRDs may account for the differential action of Rap1A on Raf-1 and B-Raf. The importance of the interaction at the CRD is further supported by a domain-shuffling experiment between Raf-1 and B-Raf, which clearly indicated that the nature of CRD determines the specificity of response to Rap1A: Raf-1, whose CRD is replaced by B-Raf CRD, became activatable by Rap1A, whereas B-Raf, whose CRD is replaced by Raf-1 CRD, lost its response to Rap1A. Finally, a B-Raf CRD mutant whose interaction with Rap1A is selectively enhanced was isolated and found to possess the double mutation K252E/M278T. B-Raf carrying this mutation was not activated by Rap1A but retained its response to Ras. These results indicate that the strength of interaction with Ras and Rap1A at its CRD may be a critical determinant of regulation of the Raf kinase activity by the Ras family small GTPases.
Figures




References
-
- Ayruch J. Insulin signal transduction through protein kinase cascades. Mol Cell Biochem. 1998;182:31–48. - PubMed
-
- Bosch E, Cherwinski H, Peterson D, McMahon M. Mutations of critical amino acids affect the biological and biochemical properties of oncogenic A-Raf and Raf-1. Oncogene. 1997;15:1021–1033. - PubMed
-
- Brtva T R, Drugan J K, Ghosh S, Terrell R S, Campbell-Burk S, Bell R M, Der C J. Two distinct Raf domains mediate interaction with Ras. J Biol Chem. 1995;270:9809–9812. - PubMed
-
- Campbell S L, Khosravi-Far R, Rossman K L, Clark G J, Der C J. Increasing complexity of Ras signaling. Oncogene. 1998;17:1395–1413. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
