RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase
- PMID: 10454574
- PMCID: PMC84588
- DOI: 10.1128/MCB.19.9.6276
RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase
Abstract
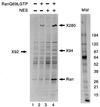
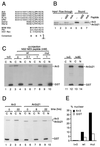
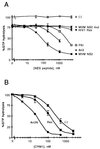
CRM1 is an export receptor mediating rapid nuclear exit of proteins and RNAs to the cytoplasm. CRM1 export cargoes include proteins with a leucine-rich nuclear export signal (NES) that bind directly to CRM1 in a trimeric complex with RanGTP. Using a quantitative CRM1-NES cargo binding assay, significant differences in affinity for CRM1 among natural NESs are demonstrated, suggesting that the steady-state nucleocytoplasmic distribution of shuttling proteins could be determined by the relative strengths of their NESs. We also show that a trimeric CRM1-NES-RanGTP complex is disassembled by RanBP1 in the presence of RanGAP, even though RanBP1 itself contains a leucine-rich NES. Selection of CRM1-binding proteins from Xenopus egg extract leads to the identification of an NES-containing DEAD-box helicase, An3, that continuously shuttles between the nucleus and the cytoplasm. In addition, we identify the Xenopus homologue of the nucleoporin CAN/Nup214 as a RanGTP- and NES cargo-specific binding site for CRM1, suggesting that this nucleoporin plays a role in export complex disassembly and/or CRM1 recycling.
Figures




References
-
- Arts G-J, Englmeier L, Mattaj I W. Energy- and temperature-dependent in vitro export of RNA from synthetic nuclei. Biol Chem. 1997;378:641–649. - PubMed
-
- Arts G-J, Fornerod M, Mattaj I W. Identification of a nuclear export receptor for tRNA. Curr Biol. 1998;8:305–314. - PubMed
-
- Askjaer P, Jensen T H, Nilsson J, Englmeier L, Kjems J. The specificity of the CRM1-Rev nuclear export signal interaction is mediated by RanGTP. J Biol Chem. 1998;273:33414–33422. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous
