Role of ornibactin biosynthesis in the virulence of Burkholderia cepacia: characterization of pvdA, the gene encoding L-ornithine N(5)-oxygenase
- PMID: 10456885
- PMCID: PMC96763
- DOI: 10.1128/IAI.67.9.4443-4455.1999
Role of ornibactin biosynthesis in the virulence of Burkholderia cepacia: characterization of pvdA, the gene encoding L-ornithine N(5)-oxygenase
Abstract
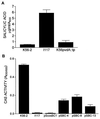
Burkholderia cepacia is a frequent cause of respiratory infections in cystic fibrosis patients. B. cepacia has been shown to produce at least four siderophores which may play a role in the virulence of this organism. To characterize genes involved in the synthesis of siderophores, Tn5-OT182 mutants were isolated in strain K56-2, which produces two siderophores, salicylic acid (SA) and ornibactins. Two mutants were characterized that did not produce zones on Chrome Azurol S agar in a commonly used assay to detect siderophore activity. These mutants were determined to produce sevenfold more SA than K56-2 yet did not produce detectable amounts of ornibactins. These mutants, designated I117 and T10, had a transposon insertion in genes with significant homology to pyoverdine biosynthesis genes of Pseudomonas aeruginosa. I117 contained an insertion in a pvdA homolog, the gene for the enzyme L-ornithine N(5)-oxygenase, which catalyzes the hydroxylation of L-ornithine. Ornibactin synthesis in this mutant was partially restored when the precursor L-N(5)-OH-Orn was added to the culture medium. T10 contained an insertion in a pvdD homolog, which is a peptide synthetase involved in pyoverdine synthesis. beta-Galactosidase activity was iron regulated in both I117 and T10, suggesting that the transposon was inserted downstream of an iron-regulated promoter. Tn5-OT182 contains a lacZ gene that is expressed when inserted downstream of an active promoter. Both I117 and T10 were deficient in uptake of iron complexed to either ornibactins or SA, suggesting that transposon insertions in ornibactin biosynthesis genes also affected other components of the iron transport mechanism. The B. cepacia pvdA homolog was approximately 47% identical and 59% similar to L-ornithine N(5)-oxygenase from P. aeruginosa. Three clones were identified from a K56-2 cosmid library that partially restored ornibactin production, SA production, and SA uptake to parental levels but did not affect the rate of (59)Fe-ornibactin uptake in I117. A chromosomal pvdA deletion mutant was constructed that had a phenotype similar to that of I117 except that it did not hyperproduce SA. The pvdA mutants were less virulent than the parent strain in chronic and acute models of respiratory infection. A functional pvdA gene appears to be required for effective colonization and persistence in B. cepacia lung infections.
Figures
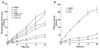

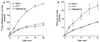
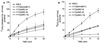

References
-
- Atkin C L, Neilands J B. Rhodotorulic acid, a diketopiperazine dihydroxamic acid with growth-factor activity. I. Isolation and characterization. Biochemistry. 1968;7:3734–3739. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1989.
-
- Braun V, Gunter K, Hantke K. Transport of iron across the outer membrane. Biol Metals. 1991;4:14–22. - PubMed
-
- Cantin, A. M., and D. E. Woods. Prolastin suppresses lung damage, inflammation and bacterial proliferation in a model of chronic Pseudomonas aeruginosa lung infection. Am. Rev. Res. Crit. Care Med., in press. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

