Kinetic differences at the single molecule level account for the functional diversity of rabbit cardiac myosin isoforms
- PMID: 10457082
- PMCID: PMC2269540
- DOI: 10.1111/j.1469-7793.1999.0669n.x
Kinetic differences at the single molecule level account for the functional diversity of rabbit cardiac myosin isoforms
Abstract
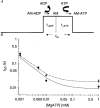
1. Cardiac V3 myosin generates slower actin filament velocities and higher average isometric forces (in an in vitro motility assay) when compared with the V1 isoform. 2. To account for differences in V1 and V3 force and motion generation at the molecular level, we characterized the mechanics and kinetics of single V1 and V3 myosin molecules using a dual laser trap setup. 3. No differences in either unitary displacement (approximately 7 nm) or force (approximately 0.8 pN) were observed between isoforms; however, the duration of unitary displacement events was significantly longer for the V3 isoform at MgATP concentrations > 10 microM. 4. Our results were interpreted on the basis of a cross-bridge model in which displacement event durations were determined by the rates of MgADP release from, and MgATP binding to, myosin. 5. We propose that the release rate of MgADP from V3 myosin is half that of V1 myosin without any difference in their rates of MgATP binding; thus, kinetic differences between the two cardiac myosin isoforms are sufficient to account for their functional diversity.
Figures


References
-
- Alousi AA, Grant AM, Etzler JR, Cofer BR, van der Bel-Kahn J, Melvin D. Reduced cardiac myofibrillar Mg-ATPase activity without changes in myosin isoenzymes in patients with end-stage failure. Molecular and Cellular Biochemistry. 1990;96:79–88. - PubMed
-
- Alpert NR, Mulieri LA. Increased myothermal economy of isometric force generation in compensated cardiac hypertrophy induced by pulmonary artery constriction in the rabbit. Circulation Research. 1982;50:491–500. - PubMed
-
- Cappelli V, Bottinelli R, Poggesi C, Moggio R, Reggiani C. Shortening velocity and myosin and myofibrillar ATPase activity related to myosin isoenzyme composition during postnatal development in rat myocardium. Circulation Research. 1989;65:446–457. - PubMed
-
- Dupuis DE, Guilford WH, Wu J, Warshaw DM. Actin filament mechanics in the laser trap. Journal of Muscle Research and Cell Motility. 1997;18:17–30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

