Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex
- PMID: 10459007
- PMCID: PMC2156148
- DOI: 10.1083/jcb.146.4.709
Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex
Abstract
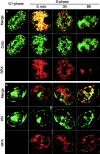
Previous experiments in Xenopus egg extracts identified what appeared to be two independently assembled prereplication complexes (pre-RCs) for DNA replication: the stepwise assembly of ORC, Cdc6, and Mcm onto chromatin, and the FFA-1-mediated recruitment of RPA into foci on chromatin. We have investigated whether both of these pre-RCs can be detected in Chinese hamster ovary (CHO) cells. Early- and late-replicating chromosomal domains were pulse-labeled with halogenated nucleotides and prelabeled cells were synchronized at various times during the following G1-phase. The recruitment of Mcm2 and RPA to these domains was examined in relation to the formation of a nuclear envelope, specification of the dihydrofolate reductase (DHFR) replication origin and entry into S-phase. Mcm2 was loaded gradually and cumulatively onto both early- and late-replicating chromatin from late telophase throughout G1-phase. During S-phase, detectable Mcm2 was rapidly excluded from PCNA-containing active replication forks. By contrast, detergent-resistant RPA foci were undetectable until the onset of S-phase, when RPA joined only the earliest-firing replicons. During S-phase, RPA was present with PCNA specifically at active replication forks. Together, our data are consistent with a role for Mcm proteins, but not RPA, in the formation of mammalian pre-RCs during early G1-phase.
Figures









Similar articles
-
Mammalian nuclei become licensed for DNA replication during late telophase.J Cell Sci. 2002 Jan 1;115(Pt 1):51-9. doi: 10.1242/jcs.115.1.51. J Cell Sci. 2002. PMID: 11801723 Free PMC article.
-
Temporally coordinated assembly and disassembly of replication factories in the absence of DNA synthesis.Nat Cell Biol. 2000 Oct;2(10):686-94. doi: 10.1038/35036309. Nat Cell Biol. 2000. PMID: 11025658 Free PMC article.
-
Stability, chromatin association and functional activity of mammalian pre-replication complex proteins during the cell cycle.EMBO J. 2001 Aug 1;20(15):4263-77. doi: 10.1093/emboj/20.15.4263. EMBO J. 2001. PMID: 11483529 Free PMC article.
-
Dynamics of pre-replication complex proteins during the cell division cycle.Philos Trans R Soc Lond B Biol Sci. 2004 Jan 29;359(1441):7-16. doi: 10.1098/rstb.2003.1360. Philos Trans R Soc Lond B Biol Sci. 2004. PMID: 15065651 Free PMC article. Review.
-
Behavior of replication origins in Eukaryota - spatio-temporal dynamics of licensing and firing.Cell Cycle. 2015;14(14):2251-64. doi: 10.1080/15384101.2015.1056421. Epub 2015 Jun 1. Cell Cycle. 2015. PMID: 26030591 Free PMC article. Review.
Cited by
-
G2 phase chromatin lacks determinants of replication timing.J Cell Biol. 2010 Jun 14;189(6):967-80. doi: 10.1083/jcb.201002002. Epub 2010 Jun 7. J Cell Biol. 2010. PMID: 20530209 Free PMC article.
-
Mammalian Orc1 protein is selectively released from chromatin and ubiquitinated during the S-to-M transition in the cell division cycle.Mol Cell Biol. 2002 Jan;22(1):105-16. doi: 10.1128/MCB.22.1.105-116.2002. Mol Cell Biol. 2002. PMID: 11739726 Free PMC article.
-
Distinct action of the retinoblastoma pathway on the DNA replication machinery defines specific roles for cyclin-dependent kinase complexes in prereplication complex assembly and S-phase progression.Mol Cell Biol. 2006 Oct;26(20):7667-81. doi: 10.1128/MCB.00045-06. Epub 2006 Aug 14. Mol Cell Biol. 2006. Retraction in: Mol Cell Biol. 2020 Aug 14;40(17):e00319-20. doi: 10.1128/MCB.00319-20. PMID: 16908528 Free PMC article. Retracted.
-
The replicative helicase MCM recruits cohesin acetyltransferase ESCO2 to mediate centromeric sister chromatid cohesion.EMBO J. 2018 Aug 1;37(15):e97150. doi: 10.15252/embj.201797150. Epub 2018 Jun 21. EMBO J. 2018. PMID: 29930102 Free PMC article.
-
FANCD2 limits acetaldehyde-induced genomic instability during DNA replication in esophageal keratinocytes.Mol Oncol. 2021 Nov;15(11):3109-3124. doi: 10.1002/1878-0261.13072. Epub 2021 Aug 8. Mol Oncol. 2021. PMID: 34328261 Free PMC article.
References
-
- Aparicio O.M., Weinstein D.M., Bell S.P. Components and dynamics of DNA replication complexes in S. cerevisiaeredistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. - PubMed
-
- Aten J.A., Bakker P.J.M., Stap J., Boschman G.A., Veenhof C.H.N. DNA double labelling with IdUrd and CldUrd for spatial and temporal analysis of cell proliferation and DNA replication. Histochem. J. 1992;24:251–259. - PubMed
-
- Bakker P.J., Stap J., Tukker C.J., van Oven C.H., Veenhof C.H., Aten J. An indirect immunofluorescence double staining procedure for the simultaneous flow cytometric measurement of iodo- and chlorodeoxyuridine incorporated into DNA. Cytometry. 1991;12:366–372. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

