The ribosome regulates the GTPase of the beta-subunit of the signal recognition particle receptor
- PMID: 10459008
- PMCID: PMC2156146
- DOI: 10.1083/jcb.146.4.723
The ribosome regulates the GTPase of the beta-subunit of the signal recognition particle receptor
Abstract
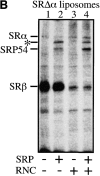
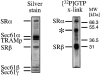
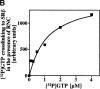
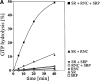
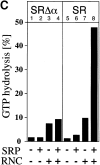
Protein targeting to the membrane of the ER is regulated by three GTPases, the 54-kD subunit of the signal recognition particle (SRP) and the alpha- and beta-subunit of the SRP receptor (SR). Here, we report on the GTPase cycle of the beta-subunits of the SR (SRbeta). We found that SRbeta binds GTP with high affinity and interacts with ribosomes in the GTP-bound state. Subsequently, the ribosome increases the GTPase activity of SRbeta and thus functions as a GTPase activating protein for SRbeta. Furthermore, the interaction between SRbeta and the ribosome leads to a reduction in the affinity of SRbeta for guanine nucleotides. We propose that SRbeta regulates the interaction of SR with the ribosome and thereby allows SRalpha to scan membrane-bound ribosomes for the presence of SRP. Interaction between SRP and SRalpha then leads to release of the signal sequence from SRP and insertion into the translocon. GTP hydrolysis then results in dissociation of SR from the ribosome, and SRP from the SR.
Figures







References
-
- Bacher G., Lütcke H., Jungnickel B., Rapoport T.A., Dobberstein B. Regulation by the ribosome of the GTPase of the signal recognition particle during protein targeting. Nature. 1996;381:248–251. - PubMed
-
- Beckmann R., Bubeck D., Grassucci R., Penczek P., Verschoor A., Blobel G., Frank J. Alignment of conduits for the nascent polypeptide chain in the ribosome–Sec61 complex. Nature. 1997;278:2123–2126. - PubMed
-
- Bernstein H.D., Poritz M.A., Strub K., Hoben P.J., Brenner S., Walter P. Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature. 1989;340:482–486. - PubMed
-
- Borgese D., Mok W., Kreibich G., Sabatini D.D. Ribosome–membrane interaction. In vitro binding of ribosomes to microsomal membranes. J. Mol. Biol. 1974;88:559–580. - PubMed
-
- Bourne H.R., Sanders D.A., McCormick F. The GTPase superfamilyconserved structure and molecular mechanism. Nature. 1991;349:117–127. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

