Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane
- PMID: 10459010
- PMCID: PMC2156145
- DOI: 10.1083/jcb.146.4.741
Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane
Abstract
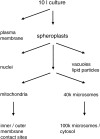
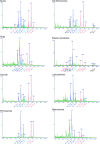
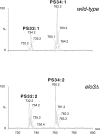
Nano-electrospray ionization tandem mass spectrometry (nano-ESI-MS/MS) was employed to determine qualitative differences in the lipid molecular species composition of a comprehensive set of organellar membranes, isolated from a single culture of Saccharomyces cerevisiae cells. Remarkable differences in the acyl chain composition of biosynthetically related phospholipid classes were observed. Acyl chain saturation was lowest in phosphatidylcholine (15.4%) and phosphatidylethanolamine (PE; 16.2%), followed by phosphatidylserine (PS; 29.4%), and highest in phosphatidylinositol (53.1%). The lipid molecular species profiles of the various membranes were generally similar, with a deviation from a calculated average profile of approximately +/- 20%. Nevertheless, clear distinctions between the molecular species profiles of different membranes were observed, suggesting that lipid sorting mechanisms are operating at the level of individual molecular species to maintain the specific lipid composition of a given membrane. Most notably, the plasma membrane is enriched in saturated species of PS and PE. The nature of the sorting mechanism that determines the lipid composition of the plasma membrane was investigated further. The accumulation of monounsaturated species of PS at the expense of diunsaturated species in the plasma membrane of wild-type cells was reversed in elo3Delta mutant cells, which synthesize C24 fatty acid-substituted sphingolipids instead of the normal C26 fatty acid-substituted species. This observation suggests that acyl chain-based sorting and/or remodeling mechanisms are operating to maintain the specific lipid molecular species composition of the yeast plasma membrane.
Figures



References
-
- Allan D., Kallen K.-J. Is plasma membrane lipid composition defined in the exocytic or the endocytic pathway? Trends Cell Biol. 1994;4:350–353. - PubMed
-
- Aris J.P., Blobel G. Isolation of yeast nuclei. Methods Enzymol. 1991;194:735–748. - PubMed
-
- Balasubramanian K., Gupta C.M. Transbilayer phosphatidylethanolamine movements in the yeast plasma membraneevidence for a protein-mediated, energy-dependent mechanism. Eur. J. Biochem. 1996;240:798–806. - PubMed
-
- Bligh E.G., Dyer W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. - PubMed
-
- Bretscher M.S., Munro S. Cholesterol and the Golgi apparatus. Science. 1993;261:1280–1281. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

