The yeast exosome and human PM-Scl are related complexes of 3' --> 5' exonucleases
- PMID: 10465791
- PMCID: PMC316947
- DOI: 10.1101/gad.13.16.2148
The yeast exosome and human PM-Scl are related complexes of 3' --> 5' exonucleases
Abstract
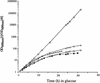
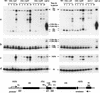
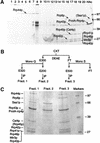
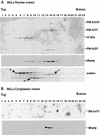
We previously identified a complex of 3' --> 5' exoribonucleases, designated the exosome, that is expected to play a major role in diverse RNA processing and degradation pathways. Further biochemical and genetic analyses have revealed six novel components of the complex. Therefore, the complex contains 11 components, 10 of which are predicted to be 3' --> 5' exoribonucleases on the basis of sequence homology. Human homologs were identified for 9 of the 11 yeast exosome components, three of which complement mutations in the respective yeast genes. Two of the newly identified exosome components are homologous to known components of the PM-Scl particle, a multisubunit complex recognized by autoimmune sera of patients suffering from polymyositis-scleroderma overlap syndrome. We demonstrate that the homolog of the Rrp4p exosome subunit is also a component of the PM-Scl complex, thereby providing compelling evidence that the yeast exosome and human PM-Scl complexes are functionally equivalent. The two complexes are similar in size, and biochemical fractionation and indirect immunofluorescence experiments show that, in both yeast and humans, nuclear and cytoplasmic forms of the complex exist that differ only by the presence of the Rrp6p/PM-Scl100 subunit exclusively in the nuclear complex.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
