Purification, molecular cloning, and expression of 2-hydroxyphytanoyl-CoA lyase, a peroxisomal thiamine pyrophosphate-dependent enzyme that catalyzes the carbon-carbon bond cleavage during alpha-oxidation of 3-methyl-branched fatty acids
- PMID: 10468558
- PMCID: PMC17838
- DOI: 10.1073/pnas.96.18.10039
Purification, molecular cloning, and expression of 2-hydroxyphytanoyl-CoA lyase, a peroxisomal thiamine pyrophosphate-dependent enzyme that catalyzes the carbon-carbon bond cleavage during alpha-oxidation of 3-methyl-branched fatty acids
Abstract
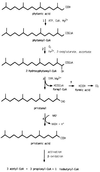
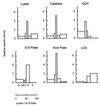
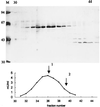
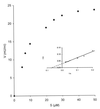
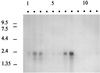
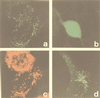
In the third step of the alpha-oxidation of 3-methyl-branched fatty acids such as phytanic acid, a 2-hydroxy-3-methylacyl-CoA is cleaved into formyl-CoA and a 2-methyl-branched fatty aldehyde. The cleavage enzyme was purified from the matrix protein fraction of rat liver peroxisomes and identified as a protein made up of four identical subunits of 63 kDa. Its activity proved to depend on Mg(2+) and thiamine pyrophosphate, a hitherto unrecognized cofactor of alpha-oxidation. Formyl-CoA and 2-methylpentadecanal were identified as reaction products when the purified enzyme was incubated with 2-hydroxy-3-methylhexadecanoyl-CoA as the substrate. Hence the enzyme catalyzes a carbon-carbon cleavage, and we propose calling it 2-hydroxyphytanoyl-CoA lyase. Sequences derived from tryptic peptides of the purified rat protein were used as queries to recover human expressed sequence tags from the databases. The composite cDNA sequence of the human lyase contained an ORF of 1,734 bases that encodes a polypeptide with a calculated molecular mass of 63,732 Da. Recombinant human protein, expressed in mammalian cells, exhibited lyase activity. The lyase displayed homology to a putative Caenorhabditis elegans protein that resembles bacterial oxalyl-CoA decarboxylases. Similarly to the decarboxylases, a thiamine pyrophosphate-binding consensus domain was present in the C-terminal part of the lyase. Although no peroxisome targeting signal, neither 1 nor 2, was apparent, transfection experiments with constructs encoding green fluorescent protein fused to the full-length lyase or its C-terminal pentapeptide indicated that the C terminus of the lyase represents a peroxisome targeting signal 1 variant.
Figures

References
-
- Steinberg D. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. New York: McGraw–Hill; 1995. pp. 2351–2369.
-
- Mihalik S J, Rainville A M, Watkins P A. Eur J Biochem. 1995;232:545–551. - PubMed
-
- Croes K, Casteels M, de Hoffmann E, Mannaerts G P, Van Veldhoven P P. Eur J Biochem. 1996;240:674–683. - PubMed
-
- Croes K, Van Veldhoven P P, Mannaerts G P, Casteels M. FEBS Lett. 1997;407:197–200. - PubMed
-
- Croes K, Casteels M, Asselberghs S, Herdewijn P, Mannaerts G P, Van Veldhoven P P. FEBS Lett. 1997;412:643–645. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

