In vivo transcription factor recruitment during thyroid hormone receptor-mediated activation
- PMID: 10468567
- PMCID: PMC17847
- DOI: 10.1073/pnas.96.18.10092
In vivo transcription factor recruitment during thyroid hormone receptor-mediated activation
Abstract
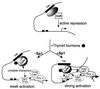
Thyroid hormone receptor (TR) can act as both a transcriptional activator and a silencer. Optimal activation by TR requires synergism with activator(s) bound to the promoter (promoter proximal activator). It is thought that liganded TR either helps to recruit preinitiation complexes (PIC) to the promoter or activates the PIC already recruited. However, the studies analyzing the TR action on the PIC formation were done in vitro and, therefore, it is not clear how relevant they are to the in vivo TR action. For example, in vivo, the TR can act from distances equal to or greater than a kilobase from the promoter, but such distant effect is not reproducible in vitro. In this study, we used the PIN*POINT (ProteIN POsition Identification with Nuclease Tail) assay to define the molecular mechanism of TR action on transcription from the thymidine kinase promoter in the cellular context. We demonstrate that the recruitment of promoter-proximal activator Sp1, and the components of the basal transcription factors such as TBP, TFIIB, and Cdk7, is enhanced with thyroid hormone activation. Our results suggest that DNA forms a loop with TR-mediated activation to accommodate interactions between the liganded TR complex and the complex formed on the promoter. We also show that Sp1 bound to the promoter is essential for the DNA looping and recruitment of basal transcription factors such as TFIIB and Cdk7 but not for recruitment of TBP. On the basis of these findings, we present a model that illustrates the molecular mechanism of TR-mediated activation in vivo.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

