Constructing primate phylogenies from ancient retrovirus sequences
- PMID: 10468595
- PMCID: PMC17875
- DOI: 10.1073/pnas.96.18.10254
Constructing primate phylogenies from ancient retrovirus sequences
Abstract
The genomes of modern humans are riddled with thousands of endogenous retroviruses (HERVs), the proviral remnants of ancient viral infections of the primate lineage. Most HERVs are nonfunctional, selectively neutral loci. This fact, coupled with their sheer abundance in primate genomes, makes HERVs ideal for exploitation as phylogenetic markers. Endogenous retroviruses (ERVs) provide phylogenetic information in two ways: (i) by comparison of integration site polymorphism and (ii) by orthologous comparison of evolving, proviral, nucleotide sequence. In this study, trees are constructed with the noncoding long terminal repeats (LTRs) of several ERV loci. Because the two LTRs of an ERV are identical at the time of integration but evolve independently, each ERV locus can provide two estimates of species phylogeny based on molecular evolution of the same ancestral sequence. Moreover, tree topology is highly sensitive to conversion events, allowing for easy detection of sequences involved in recombination as well as correction for such events. Although other animal species are rich in ERV sequences, the specific use of HERVs in this study allows comparison of trees to a well established phylogenetic standard, that of the Old World primates. HERVs, and by extension the ERVs of other species, constitute a unique and plentiful resource for studying the evolutionary history of the Retroviridae and their animal hosts.
Figures
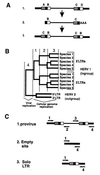

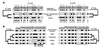
References
-
- Boeke J D, Stoye J P. In: Retroviruses. Coffin J M, Hughes S H, Varmus H E, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 343–436. - PubMed
-
- Coffin J M. In: RNA Tumor Viruses. Weiss R, Teich N, Varmus H, Coffin J M, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. pp. 1109–1204.
-
- Coffin J M. In: Fundamental Virology. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott-Raven; 1996. pp. 763–844.
-
- Benveniste R E, Todaro G J. Nature (London) 1976;261:101–108. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

