Phylogenetic relationships among cetartiodactyls based on insertions of short and long interpersed elements: hippopotamuses are the closest extant relatives of whales
- PMID: 10468596
- PMCID: PMC17876
- DOI: 10.1073/pnas.96.18.10261
Phylogenetic relationships among cetartiodactyls based on insertions of short and long interpersed elements: hippopotamuses are the closest extant relatives of whales
Abstract
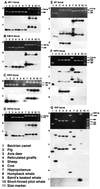
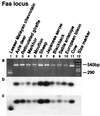
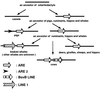
Insertion analysis of short and long interspersed elements is a powerful method for phylogenetic inference. In a previous study of short interspersed element data, it was found that cetaceans, hippopotamuses, and ruminants form a monophyletic group. To further resolve the relationships among these taxa, we now have isolated and characterized 10 additional loci. A phylogenetic analysis of these data was able to resolve relationships among the major cetartiodactyl groups, thereby shedding light on the origin of whales. The results indicated (i) that cetaceans are deeply nested within Artiodactyla, (ii) that cetaceans and hippopotamuses form a monophyletic group, (iii) that pigs and peccaries form a monophyletic group to the exclusion of hippopotamuses, (iv) that chevrotains diverged first among ruminants, and (v) that camels diverged first among cetartiodactyls. These findings lead us to conclude that cetaceans evolved from an immediate artiodactyl, not mesonychian, ancestor.
Figures




Comment in
-
SINEs of the perfect character.Proc Natl Acad Sci U S A. 1999 Aug 31;96(18):9979-81. doi: 10.1073/pnas.96.18.9979. Proc Natl Acad Sci U S A. 1999. PMID: 10468541 Free PMC article. Review. No abstract available.
References
-
- Gingerich P D, Smith B H, Simons E L. Science. 1990;249:154–157. - PubMed
-
- Thewissen J G M, Hussain S T. Nature (London) 1993;361:444–445. - PubMed
-
- Thewissen J G M. J Mammal Evol. 1994;2:157–184.
-
- Fordyce R E, Barnes L G. Annu Rev Earth Planet Sci. 1994;22:419–455.
-
- Prothero D R, Manning E M, Fischer M. In: The Phylogeny and Classification of the Tetrapods. Benton M J, editor. Vol. 2. Oxford: Clarendon; 1988. pp. 201–234.
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

