Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the alpha-subunit of GTP-binding protein
- PMID: 10468600
- PMCID: PMC17880
- DOI: 10.1073/pnas.96.18.10284
Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the alpha-subunit of GTP-binding protein
Abstract
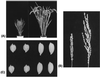
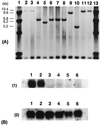
A rice Dwarf 1 gene was identified by using a map-based cloning strategy. Its recessive mutant allele confers a dwarf phenotype. Linkage analysis revealed that a cDNA encoding the alpha-subunit of GTP-binding protein cosegregated with d1 in 3,185 d1 segregants. Southern hybridization analysis with this cDNA as a probe showed different band patterns in several d1 mutant lines. In at least four independent d1 mutants, no gene transcript was observed by Northern hybridization analysis. Sequencing analysis revealed that an 833-bp deletion had occurred in one of the mutant alleles, which resulted in an inability to express GTP-binding protein. A transgenic d1 mutant with GTP-binding protein gene restored the normal phenotype. We conclude that the rice Dwarf 1 gene encodes GTP-binding protein and that the protein plays an important role in plant growth and development. Because the d1 mutant is classified as gibberellin-insensitive, we suggest that the GTP-binding protein might be associated with gibberellin signal transduction.
Figures
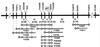

References
LinkOut - more resources
Full Text Sources
Other Literature Sources

