The AtCAO gene, encoding chlorophyll a oxygenase, is required for chlorophyll b synthesis in Arabidopsis thaliana
- PMID: 10468639
- PMCID: PMC17919
- DOI: 10.1073/pnas.96.18.10507
The AtCAO gene, encoding chlorophyll a oxygenase, is required for chlorophyll b synthesis in Arabidopsis thaliana
Abstract
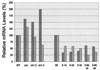
Chlorophyll b is synthesized from chlorophyll a and is found in the light-harvesting complexes of prochlorophytes, green algae, and both nonvascular and vascular plants. We have used conserved motifs from the chlorophyll a oxygenase (CAO) gene from Chlamydomonas reinhardtii to isolate a homologue from Arabidopsis thaliana. This gene, AtCAO, is mutated in both leaky and null chlorina1 alleles, and DNA sequence changes cosegregate with the mutant phenotype. AtCAO mRNA levels are higher in three different mutants that have reduced levels of chlorophyll b, suggesting that plants that do not have sufficient chlorophyll b up-regulate AtCAO gene expression. Additionally, AtCAO mRNA levels decrease in plants that are grown under dim-light conditions. We have also found that the six major Lhcb proteins do not accumulate in the null ch1-3 allele.
Figures

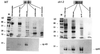

References
-
- Simpson D J, Machold O, Hoyer-Hansen G, von Wettstein D. Carlsberg Res Comm. 1985;50:223–238.
-
- Apel K, Kloppstech K. Planta. 1980;150:426–430. - PubMed
-
- Bellemare F, Bartlett S G, Chua N-H. J Biol Chem. 1982;257:7762–7767. - PubMed
-
- Kuttkat A, Edhofer I, Eichacker L A, Paulsen H. J Biol Chem. 1997;272:20451–20455. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

