Transformation of sulfur compounds by an abundant lineage of marine bacteria in the alpha-subclass of the class Proteobacteria
- PMID: 10473380
- PMCID: PMC99705
- DOI: 10.1128/AEM.65.9.3810-3819.1999
Transformation of sulfur compounds by an abundant lineage of marine bacteria in the alpha-subclass of the class Proteobacteria
Abstract
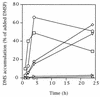
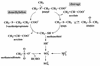
Members of a group of marine bacteria that is numerically important in coastal seawater and sediments were characterized with respect to their ability to transform organic and inorganic sulfur compounds. Fifteen strains representing the Roseobacter group (a phylogenetic cluster of marine bacteria in the alpha-subclass of the class Proteobacteria) were isolated from seawater, primarily from the southeastern United States. Although more than one-half of the isolates were obtained without any selection for sulfur metabolism, all of the isolates were able to degrade the sulfur-containing osmolyte dimethyl sulfoniopropionate (DMSP) with production of dimethyl sulfide (DMS). Five isolates also degraded DMSP with production of methanethiol, indicating that both cleavage and demethylation pathways for DMSP occurred in the same organism, which is unusual. Five isolates were able to reduce dimethyl sulfoxide to DMS, and several isolates also degraded DMS and methanethiol. Sulfite oxygenase activity and methanesulfonic acid oxygenase activity were also present in some of the isolates. The ability to incorporate the reduced sulfur in DMSP and methanethiol into cellular material was studied with one of the isolates. A group-specific 16S rRNA probe indicated that the relative abundance of uncultured bacteria in the Roseobacter group increased in seawater enriched with DMSP or DMS. Because this group typically accounts for >10% of the 16S ribosomal DNA pool in coastal seawater and sediments of the southern United States, clues about its potential biogeochemical role are of particular interest. Studies of culturable representatives suggested that the group could mediate a number of steps in the cycling of both organic and inorganic forms of sulfur in marine environments.
Figures


References
-
- Baumann P, Baumann L. The marine Gram-negative eubacteria: genera Photobacterium, Beneckea, Alteromonas, Pseudomonas, and Alcaligenes. In: Starr M P, Stolp H, Trüper H G, Balows A, Schlegel H G, editors. The prokaryotes. Berlin, Germany: Springer-Verlag; 1981. pp. 1302–1331.
-
- Charlson R J, Lovelock J E, Andreae M O, Warren S G. Oceanic phytoplankton, atmospheric sulfur, cloud albedo and climate. Nature (London) 1987;326:655–661.
-
- Dacey J W H, Blough N V. Hydroxide decomposition of dimethylsulfoniopropionate to form dimethylsulfide. Geophys Res Lett. 1987;14:1246–1249.
-
- DeLong E F, Franks D G, Alldredge A L. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol Oceanogr. 1993;38:924–934.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

