Cloning of the gene encoding a novel thermostable alpha-galactosidase from Thermus brockianus ITI360
- PMID: 10473401
- PMCID: PMC99726
- DOI: 10.1128/AEM.65.9.3955-3963.1999
Cloning of the gene encoding a novel thermostable alpha-galactosidase from Thermus brockianus ITI360
Abstract
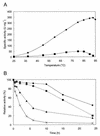
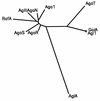
An alpha-galactosidase gene from Thermus brockianus ITI360 was cloned, sequenced, and expressed in Escherichia coli, and the recombinant protein was purified. The gene, designated agaT, codes for a 476-residue polypeptide with a calculated molecular mass of 53, 810 Da. The native structure of the recombinant enzyme (AgaT) was estimated to be a tetramer. AgaT displays amino acid sequence similarity to the alpha-galactosidases of Thermotoga neapolitana and Thermotoga maritima and a low-level sequence similarity to alpha-galactosidases of family 36 in the classification of glycosyl hydrolases. The enzyme is thermostable, with a temperature optimum of activity at 93 degrees C with para-nitrophenyl-alpha-galactopyranoside as a substrate. Half-lives of inactivation at 92 and 80 degrees C are 100 min and 17 h, respectively. The pH optimum is between 5.5 and 6.5. The enzyme displayed high affinity for oligomeric substrates. The K(m)s for melibiose and raffinose at 80 degrees C were determined as 4.1 and 11.0 mM, respectively. The alpha-galactosidase gene in T. brockianus ITI360 was inactivated by integrational mutagenesis. Consequently, no alpha-galactosidase activity was detectable in crude extracts of the mutant strain, and it was unable to use melibiose or raffinose as a single carbohydrate source.
Figures





Similar articles
-
The structure of the alpha-galactosidase gene loci in Thermus brockianus ITI360 and Thermus thermophilus TH125.Extremophiles. 2000 Feb;4(1):23-33. doi: 10.1007/s007920050004. Extremophiles. 2000. PMID: 10741834
-
Purification and characterization of the recombinant Thermus sp. strain T2 alpha-galactosidase expressed in Escherichia coli.Appl Environ Microbiol. 2001 Apr;67(4):1601-6. doi: 10.1128/AEM.67.4.1601-1616.2001. Appl Environ Microbiol. 2001. PMID: 11282611 Free PMC article.
-
Properties of an alpha-galactosidase, and structure of its gene galA, within an alpha-and beta-galactoside utilization gene cluster of the hyperthermophilic bacterium Thermotoga maritima.Syst Appl Microbiol. 1998 Mar;21(1):1-11. doi: 10.1016/s0723-2020(98)80002-7. Syst Appl Microbiol. 1998. PMID: 9741105
-
Thermostable alpha-galactosidase from Bacillus stearothermophilus NUB3621: cloning, sequencing and characterization.FEMS Microbiol Lett. 1999 Jul 1;176(1):147-53. doi: 10.1111/j.1574-6968.1999.tb13655.x. FEMS Microbiol Lett. 1999. PMID: 10418141
-
Thermostable alpha-galactosidase from Thermotoga neapolitana: cloning, sequencing and expression.FEMS Microbiol Lett. 1998 Jun 1;163(1):37-42. doi: 10.1111/j.1574-6968.1998.tb13023.x. FEMS Microbiol Lett. 1998. PMID: 9631543
Cited by
-
Partial Characterization of α-Galactosidic Activity from the Antarctic Bacterial Isolate, Paenibacillus sp. LX-20 as a Potential Feed Enzyme Source.Asian-Australas J Anim Sci. 2012 Jun;25(6):852-60. doi: 10.5713/ajas.2011.11501. Asian-Australas J Anim Sci. 2012. PMID: 25049637 Free PMC article.
-
Identification by saturation mutagenesis of a single residue involved in the alpha-galactosidase AgaB regioselectivity.Glycoconj J. 2001 Jun;18(6):457-64. doi: 10.1023/a:1016034101436. Glycoconj J. 2001. PMID: 12084981
-
Biodegradation of Polymers Used in Oil and Gas Operations: Towards Enzyme Biotechnology Development and Field Application.Polymers (Basel). 2022 May 3;14(9):1871. doi: 10.3390/polym14091871. Polymers (Basel). 2022. PMID: 35567040 Free PMC article. Review.
-
Thermoadaptation of alpha-galactosidase AgaB1 in Thermus thermophilus.J Bacteriol. 2002 Jun;184(12):3385-91. doi: 10.1128/JB.184.12.3385-3391.2002. J Bacteriol. 2002. PMID: 12029056 Free PMC article.
-
Characterization of Properties and Transglycosylation Abilities of Recombinant α-Galactosidase from Cold-Adapted Marine Bacterium Pseudoalteromonas KMM 701 and Its C494N and D451A Mutants.Mar Drugs. 2018 Sep 24;16(10):349. doi: 10.3390/md16100349. Mar Drugs. 2018. PMID: 30250010 Free PMC article.
References
-
- Aduse-Opoku J, Tao L, Ferretti J J, Russell R R B. Biochemical and genetic analysis of Streptococcus mutans α-galactosidase. J Gen Microbiol. 1991;137:757–764. - PubMed
-
- Altenbuchner J. A new λ RES vector with a built-in Tn1721-encoded excision system. Gene. 1993;123:63–68. - PubMed
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Berger J L, Lee L H, Lacroix C. Identification of new enzyme activities of several strains of Thermus species. Appl Microbiol Biotechnol. 1995;44:81–87. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

