Comparison of bacterial community structures in the rhizoplane of tomato plants grown in soils suppressive and conducive towards bacterial wilt
- PMID: 10473407
- PMCID: PMC99732
- DOI: 10.1128/AEM.65.9.3996-4001.1999
Comparison of bacterial community structures in the rhizoplane of tomato plants grown in soils suppressive and conducive towards bacterial wilt
Abstract
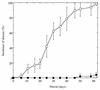
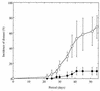
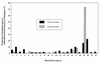
It has been reported that the growth of Ralstonia solanacearum is suppressed at the rhizoplane of tomato plants and that tomato bacterial wilt is suppressed in plants grown in a soil (Mutsumi) in Japan. To evaluate the biological factors contributing to the suppressiveness of the soil in three treated Mutsumi soils (chloroform fumigated soil; autoclaved soil mixed with intact Mutsumi soil; and autoclaved soil mixed with intact, wilt-conducive Yamadai soil) infested with R. solanacearum, we bioassayed soil samples for tomato bacterial wilt. Chloroform fumigation increased the extent of wilt disease. More of the tomato plant samples wilted when mixed with Yamadai soil than when mixed with Mutsumi soil. Consequently, the results indicate that the naturally existing population of microorganisms in Mutsumi soil was significantly able to reduce the severity of bacterial wilt of tomato plants. To characterize the types of bacteria present at the rhizoplane, we isolated rhizoplane bacteria and classified them into 22 groups by comparing their 16S restriction fragment length polymorphism patterns. In Yamadai soil a single group of bacteria was extremely predominant (73.1%), whereas in Mutsumi soil the distribution of the bacterial groups was much more even. The 16S rDNA sequence analysis of strains of dominant groups suggested that gram-negative bacteria close to the beta-proteobacteria were most common at the rhizoplane of the tomato plants. During in vitro assays, rhizoplane bacteria in Mutsumi soil grew more vigorously on pectin, one of the main root exudates of tomato, compared with those in Yamadai soil. Our results imply that it is difficult for the pathogen to dominate in a diversified rhizobacterial community that thrives on pectin.
Figures

References
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Drew M C. Root function, development, growth and mineral nutrition. In: Lynch J M, editor. The rhizosphere. New York, N.Y: Wiley Interscience; 1989. pp. 35–57.
-
- Guevara M A, Gonzalez-Jaen M T, Estevez P. Multiple forms of pectic lyase and polygalacturonase from Fusarium oxysporum f.sp. radicislycopersici: regulation of their synthesis by galacturonic acid. Can J Microbiol. 1997;43:245–253.
-
- Hara H, Ono K. Ecological study on the bacterial wilt of tobacco, caused by Pseudomonas solanacearum. Bull Okayama Tob Exp Stn. 1983;42:127–138.
-
- Hartman G L, Elphinstone J G. Advance in the control of Pseudomonas solanacearum race 1 in a major food crop. In: Hayward A C, Hartman G L, editors. Bacterial wilt, the disease and its causative agent, Pseudomonas solanacearum. Wallingford, United Kingdom: CAB International; 1994. pp. 157–177.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

