Identification of some of the major groups of bacteria in efficient and nonefficient biological phosphorus removal activated sludge systems
- PMID: 10473419
- PMCID: PMC99744
- DOI: 10.1128/AEM.65.9.4077-4084.1999
Identification of some of the major groups of bacteria in efficient and nonefficient biological phosphorus removal activated sludge systems
Abstract
To investigate the bacteria that are important to phosphorus (P) removal in activated sludge, microbial populations were analyzed during the operation of a laboratory-scale reactor with various P removal performances. The bacterial population structure, analyzed by fluorescence in situ hybridization (FISH) with oligonucleotides probes complementary to regions of the 16S and 23S rRNAs, was associated with the P removal performance of the reactor. At one stage of the reactor operation, chemical characterization revealed that extremely poor P removal was occurring. However, like in typical P-removing sludges, complete anaerobic uptake of the carbon substrate occurred. Bacteria inhibiting P removal overwhelmed the reactor, and according to FISH, bacteria of the beta subclass of the class Proteobacteria other than beta-1 or beta-2 were dominant in the sludge (58% of the population). Changes made to the operation of the reactor led to the development of a biomass population with an extremely good P removal capacity. The biochemical transformations observed in this sludge were characteristic of typical P-removing activated sludge. The microbial population analysis of the P-removing sludge indicated that bacteria of the beta-2 subclass of the class Proteobacteria and actinobacteria were dominant (55 and 35%, respectively), therefore implicating bacteria from these groups in high-performance P removal. The changes in operation that led to the improved performance of the reactor included allowing the pH to rise during the anaerobic period, which promoted anaerobic phosphate release and possibly caused selection against non-phosphate-removing bacteria.
Figures
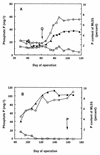

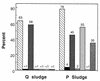
 ), BONE23a (▧),
BTWO23a (▨), GAM42a (□), and HGC69a
(
), BONE23a (▧),
BTWO23a (▨), GAM42a (□), and HGC69a
( ) are
expressed as percentages of the number of cells detected with the DAPI
stain.
) are
expressed as percentages of the number of cells detected with the DAPI
stain.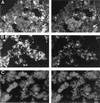
References
-
- American Public Health Association, American Water Works Association, and Water Pollution Control Federation. Standard methods for the examination of water and wastewater. 18th ed. Baltimore, Md: Port City Press; 1992.
-
- Appeldoorn L J, Kortstee G, Zehnder A J B. Biological phosphate removal by activated sludge under defined conditions. Water Res. 1992;26:453–460.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

