Molecular and biochemical characterization of two xylanase-encoding genes from Cellulomonas pachnodae
- PMID: 10473422
- PMCID: PMC99747
- DOI: 10.1128/AEM.65.9.4099-4107.1999
Molecular and biochemical characterization of two xylanase-encoding genes from Cellulomonas pachnodae
Abstract
Two xylanase-encoding genes, named xyn11A and xyn10B, were isolated from a genomic library of Cellulomonas pachnodae by expression in Escherichia coli. The deduced polypeptide, Xyn11A, consists of 335 amino acids with a calculated molecular mass of 34,383 Da. Different domains could be identified in the Xyn11A protein on the basis of homology searches. Xyn11A contains a catalytic domain belonging to family 11 glycosyl hydrolases and a C-terminal xylan binding domain, which are separated from the catalytic domain by a typical linker sequence. Binding studies with native Xyn11A and a truncated derivative of Xyn11A, lacking the putative binding domain, confirmed the function of the two domains. The second xylanase, designated Xyn10B, consists of 1,183 amino acids with a calculated molecular mass of 124,136 Da. Xyn10B also appears to be a modular protein, but typical linker sequences that separate the different domains were not identified. It comprises a N-terminal signal peptide followed by a stretch of amino acids that shows homology to thermostabilizing domains. Downstream of the latter domain, a catalytic domain specific for family 10 glycosyl hydrolases was identified. A truncated derivative of Xyn10B bound tightly to Avicel, which was in accordance with the identified cellulose binding domain at the C terminus of Xyn10B on the basis of homology. C. pachnodae, a (hemi)cellulolytic bacterium that was isolated from the hindgut of herbivorous Pachnoda marginata larvae, secretes at least two xylanases in the culture fluid. Although both Xyn11A and Xyn10B had the highest homology to xylanases from Cellulomonas fimi, distinct differences in the molecular organizations of the xylanases from the two Cellulomonas species were identified.
Figures





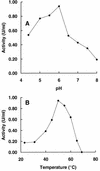
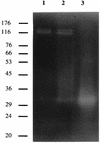
References
-
- Bayon C, Mathelin J. Carbohydrate fermentation and by-product adsorption studied with labelled cellulose in Oryctes nasicornis larvae (Coleoptera: Scrarabaeidae) J Insect Physiol. 1980;26:833–840.
-
- Bhalerao J, Patki A H, Bhave M, Khurana I, Deobagkar D N. Molecular cloning and expression of a xylanase gene from Cellulomonas sp. into Escherichia coli. Appl Microbiol Biotechnol. 1990;34:71–76.
-
- Cazemier A E, Op den Camp H J M, Hackstein J H P, Vogels G D. Fibre digestion in arthropods. Comp Biochem Physiol. 1997;118A:101–109.
-
- Cazemier, A. E., H. J. M. Op den Camp, J. C. Verdoes, J. H. P. Hackstein, and G. D. Vogels. Cellulomonas pachnodae sp. nov., a member of the (hemi)cellulolytic hindgut flora of larvae of the scarab beetle Pachnoda marginata. Submitted for publication. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

