Identification of nitrite-oxidizing bacteria with monoclonal antibodies recognizing the nitrite oxidoreductase
- PMID: 10473425
- PMCID: PMC99750
- DOI: 10.1128/AEM.65.9.4126-4133.1999
Identification of nitrite-oxidizing bacteria with monoclonal antibodies recognizing the nitrite oxidoreductase
Abstract
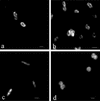
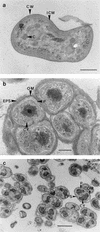
Immunoblot analyses performed with three monoclonal antibodies (MAbs) that recognized the nitrite oxidoreductase (NOR) of the genus Nitrobacter were used for taxonomic investigations of nitrite oxidizers. We found that these MAbs were able to detect the nitrite-oxidizing systems (NOS) of the genera Nitrospira, Nitrococcus, and Nitrospina. The MAb designated Hyb 153-2, which recognized the alpha subunit of the NOR (alpha-NOR), was specific for species belonging to the genus Nitrobacter. In contrast, Hyb 153-3, which recognized the beta-NOR, reacted with nitrite oxidizers of the four genera. Hyb 153-1, which also recognized the beta-NOR, bound to members of the genera Nitrobacter and Nitrococcus. The molecular masses of the beta-NOR of the genus Nitrobacter and the beta subunit of the NOS (beta-NOS) of the genus Nitrococcus were identical (65 kDa). In contrast, the molecular masses of the beta-NOS of the genera Nitrospina and Nitrospira were different (48 and 46 kDa). When the genus-specific reactions of the MAbs were correlated with 16S rRNA sequences, they reflected the phylogenetic relationships among the nitrite oxidizers. The specific reactions of the MAbs allowed us to classify novel isolates and nitrite oxidizers in enrichment cultures at the genus level. In ecological studies the immunoblot analyses demonstrated that Nitrobacter or Nitrospira cells could be enriched from activated sludge by using various substrate concentrations. Fluorescence in situ hybridization and electron microscopic analyses confirmed these results. Permeated cells of pure cultures of members of the four genera were suitable for immunofluorescence labeling; these cells exhibited fluorescence signals that were consistent with the location of the NOS.
Figures




References
-
- Beimfohr C, Krause A, Amann R, Ludwig W, Schleifer K-H. In situ identification of lactococci, enterococci and streptococci. Syst Appl Microbiol. 1993;16:450–456.
-
- Belser L W. Population ecology of nitrifying bacteria. Annu Rev Microbiol. 1979;33:309–333. - PubMed
-
- Bock E, Koops H-P. The genus Nitrobacter and related genera. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. New York, N.Y: Springer-Verlag; 1992. pp. 2302–2309.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

