A role for the GSG domain in localizing Sam68 to novel nuclear structures in cancer cell lines
- PMID: 10473643
- PMCID: PMC25546
- DOI: 10.1091/mbc.10.9.3015
A role for the GSG domain in localizing Sam68 to novel nuclear structures in cancer cell lines
Abstract
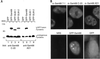
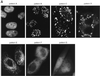
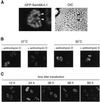
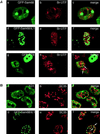
The GSG (GRP33, Sam68, GLD-1) domain is a protein module found in an expanding family of RNA-binding proteins. The numerous missense mutations identified genetically in the GSG domain support its physiological role. Although the exact function of the GSG domain is not known, it has been shown to be required for RNA binding and oligomerization. Here it is shown that the Sam68 GSG domain plays a role in protein localization. We show that Sam68 concentrates into novel nuclear structures that are predominantly found in transformed cells. These Sam68 nuclear bodies (SNBs) are distinct from coiled bodies, gems, and promyelocytic nuclear bodies. Electron microscopic studies show that SNBs are distinct structures that are enriched in phosphorus and nitrogen, indicating the presence of nucleic acids. A GFP-Sam68 fusion protein had a similar localization as endogenous Sam68 in HeLa cells, diffusely nuclear with two to five SNBs. Two other GSG proteins, the Sam68-like mammalian proteins SLM-1 and SLM-2, colocalized with endogenous Sam68 in SNBs. Different GSG domain missense mutations were investigated for Sam68 protein localization. Six separate classes of cellular patterns were obtained, including exclusive SNB localization and association with microtubules. These findings demonstrate that the GSG domain is involved in protein localization and define a new compartment for Sam68, SLM-1, and SLM-2 in cancer cell lines.
Figures





References
-
- Baehrecke EH. who encodes a KH RNA binding protein that functions in muscle development. Development. 1997;124:1323–1332. - PubMed
-
- Barlat I, Maurier F, Duchesne M, Guitard E, Tocque B, Schweighoffer F. A role for Sam68 in cell cycle progression antagonized by a spliced variant within the KH domain. J Biol Chem. 1997;272:3129–3132. - PubMed
-
- Bazett-Jones DP, Hendzel MJ. Electron spectroscopic imaging of chromatin. Methods. 1999;17:188–200. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

