The conserved core of a human SIR2 homologue functions in yeast silencing
- PMID: 10473645
- PMCID: PMC25551
- DOI: 10.1091/mbc.10.9.3045
The conserved core of a human SIR2 homologue functions in yeast silencing
Abstract
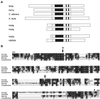
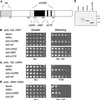
Silencing is a universal form of transcriptional regulation in which regions of the genome are reversibly inactivated by changes in chromatin structure. Sir2 (Silent Information Regulator) protein is unique among the silencing factors in Saccharomyces cerevisiae because it silences the rDNA as well as the silent mating-type loci and telomeres. Discovery of a gene family of Homologues of Sir Two (HSTs) in organisms from bacteria to humans suggests that SIR2's silencing mechanism might be conserved. The Sir2 and Hst proteins share a core domain, which includes two diagnostic sequence motifs of unknown function as well as four cysteines of a putative zinc finger. We demonstrate by mutational analyses that the conserved core and each of its motifs are essential for Sir2p silencing. Chimeras between Sir2p and a human Sir2 homologue (hSir2Ap) indicate that this human protein's core can substitute for that of Sir2p, implicating the core as a silencing domain. Immunofluorescence studies reveal partially disrupted localization, accounting for the yeast-human chimeras' ability to function at only a subset of Sir2p's target loci. Together, these results support a model for the involvement of distinct Sir2p-containing complexes in HM/telomeric and rDNA silencing and that HST family members, including the widely expressed hSir2A, may perform evolutionarily conserved functions.
Figures






Similar articles
-
Locus specificity determinants in the multifunctional yeast silencing protein Sir2.EMBO J. 2000 Jun 1;19(11):2641-51. doi: 10.1093/emboj/19.11.2641. EMBO J. 2000. PMID: 10835361 Free PMC article.
-
A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors.Mol Cell Biol. 1999 Apr;19(4):3184-97. doi: 10.1128/MCB.19.4.3184. Mol Cell Biol. 1999. PMID: 10082585 Free PMC article.
-
Analysis of Sir2p domains required for rDNA and telomeric silencing in Saccharomyces cerevisiae.Genetics. 2000 Mar;154(3):1069-83. doi: 10.1093/genetics/154.3.1069. Genetics. 2000. PMID: 10757754 Free PMC article.
-
Sound silencing: the Sir2 protein and cellular senescence.Bioessays. 2001 Apr;23(4):327-32. doi: 10.1002/bies.1047. Bioessays. 2001. PMID: 11268038 Review.
-
Nuclear organization and silencing: trafficking of Sir proteins.Novartis Found Symp. 1998;214:114-26; discussion 126-32. doi: 10.1002/9780470515501.ch7. Novartis Found Symp. 1998. PMID: 9601014 Review.
Cited by
-
A novel allele of SIR2 reveals a heritable intermediate state of gene silencing.Genetics. 2021 May 17;218(1):iyab041. doi: 10.1093/genetics/iyab041. Genetics. 2021. PMID: 33713126 Free PMC article.
-
Sirtuin 1 and sirtuin 3: physiological modulators of metabolism.Physiol Rev. 2012 Jul;92(3):1479-514. doi: 10.1152/physrev.00022.2011. Physiol Rev. 2012. PMID: 22811431 Free PMC article. Review.
-
SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria.Proc Natl Acad Sci U S A. 2002 Oct 15;99(21):13653-8. doi: 10.1073/pnas.222538099. Epub 2002 Oct 8. Proc Natl Acad Sci U S A. 2002. PMID: 12374852 Free PMC article.
-
Nicotinamide mononucleotide attenuates glucocorticoid‑induced osteogenic inhibition by regulating the SIRT1/PGC‑1α signaling pathway.Mol Med Rep. 2020 Jul;22(1):145-154. doi: 10.3892/mmr.2020.11116. Epub 2020 May 4. Mol Med Rep. 2020. PMID: 32377728 Free PMC article.
-
Protective effects and mechanisms of sirtuins in the nervous system.Prog Neurobiol. 2011 Nov;95(3):373-95. doi: 10.1016/j.pneurobio.2011.09.001. Epub 2011 Sep 10. Prog Neurobiol. 2011. PMID: 21930182 Free PMC article. Review.
References
-
- Aparicio OM, Billington BL, Gottschling DE. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. - PubMed
-
- Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, Boeke JD. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression and chromosome stability. Genes & Dev. 1995;9:2888–2902. - PubMed
-
- Braunstein M, Rose AB, Holmes SG, Allis CD, Broach JR. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes & Dev. 1993;7:592–604. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

