Transposon Tn21, flagship of the floating genome
- PMID: 10477306
- PMCID: PMC103744
- DOI: 10.1128/MMBR.63.3.507-522.1999
Transposon Tn21, flagship of the floating genome
Abstract
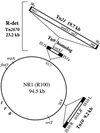
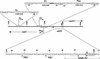
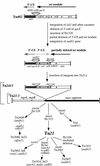
The transposon Tn21 and a group of closely related transposons (the Tn21 family) are involved in the global dissemination of antibiotic resistance determinants in gram-negative facultative bacteria. The molecular basis for their involvement is carriage by the Tn21 family of a mobile DNA element (the integron) encoding a site-specific system for the acquisition of multiple antibiotic resistance genes. The paradigm example, Tn21, also carries genes for its own transposition and a mercury resistance (mer) operon. We have compiled the entire 19,671-bp sequence of Tn21 and assessed the possible origins and functions of the genes it contains. Our assessment adds molecular detail to previous models of the evolution of Tn21 and is consistent with the insertion of the integron In2 into an ancestral Tn501-like mer transposon. Codon usage analysis indicates distinct host origins for the ancestral mer operon, the integron, and the gene cassette and two insertion sequences which lie within the integron. The sole gene of unknown function in the integron, orf5, resembles a puromycin-modifying enzyme from an antibiotic producing bacterium. A possible seventh gene in the mer operon (merE), perhaps with a role in Hg(II) transport, lies in the junction between the integron and the mer operon. Analysis of the region interrupted by insertion of the integron suggests that the putative transposition regulator, tnpM, is the C-terminal vestige of a tyrosine kinase sensor present in the ancestral mer transposon. The extensive dissemination of the Tn21 family may have resulted from the fortuitous association of a genetic element for accumulating multiple antibiotic resistances (the integron) with one conferring resistance to a toxic metal at a time when clinical, agricultural, and industrial practices were rapidly increasing the exposure to both types of selective agents. The compendium offered here will provide a reference point for ongoing observations of related elements in multiply resistant strains emerging worldwide.
Figures


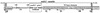
Similar articles
-
Four genes, two ends, and a res region are involved in transposition of Tn5053: a paradigm for a novel family of transposons carrying either a mer operon or an integron.Mol Microbiol. 1995 Sep;17(6):1189-200. doi: 10.1111/j.1365-2958.1995.mmi_17061189.x. Mol Microbiol. 1995. PMID: 8594337
-
Tn5053, a mercury resistance transposon with integron's ends.J Mol Biol. 1993 Apr 20;230(4):1103-7. doi: 10.1006/jmbi.1993.1228. J Mol Biol. 1993. PMID: 8387603
-
[Tn5037-a Tn21-like mercury transposon, detected in Thiobacillus ferrooxidans].Genetika. 2001 Aug;37(8):1160-4. Genetika. 2001. PMID: 11642118 Russian.
-
Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon.FEMS Microbiol Rev. 1997 Apr;19(4):239-62. doi: 10.1111/j.1574-6976.1997.tb00300.x. FEMS Microbiol Rev. 1997. PMID: 9167257 Review.
-
Site-specific recombination and shuffling of resistance genes in transposon Tn21.Res Microbiol. 1991 Jul-Aug;142(6):701-4. doi: 10.1016/0923-2508(91)90083-m. Res Microbiol. 1991. PMID: 1660178 Review.
Cited by
-
The mosaic architecture of Aeromonas salmonicida subsp. salmonicida pAsa4 plasmid and its consequences on antibiotic resistance.PeerJ. 2016 Oct 27;4:e2595. doi: 10.7717/peerj.2595. eCollection 2016. PeerJ. 2016. PMID: 27812409 Free PMC article.
-
Novel macrolide resistance module carried by the IncP-1beta resistance plasmid pRSB111, isolated from a wastewater treatment plant.Antimicrob Agents Chemother. 2007 Feb;51(2):673-8. doi: 10.1128/AAC.00802-06. Epub 2006 Nov 13. Antimicrob Agents Chemother. 2007. PMID: 17101677 Free PMC article.
-
Recruitment of Mobile Genetic Elements for Diverse Cellular Functions in Prokaryotes.Front Mol Biosci. 2022 Mar 24;9:821197. doi: 10.3389/fmolb.2022.821197. eCollection 2022. Front Mol Biosci. 2022. PMID: 35402511 Free PMC article. Review.
-
DNA sequence and comparative genomics of pAPEC-O2-R, an avian pathogenic Escherichia coli transmissible R plasmid.Antimicrob Agents Chemother. 2005 Nov;49(11):4681-8. doi: 10.1128/AAC.49.11.4681-4688.2005. Antimicrob Agents Chemother. 2005. PMID: 16251312 Free PMC article.
-
Emergence of multidrug-resistant Salmonella enterica serovar typhi in Korea.Antimicrob Agents Chemother. 2004 Nov;48(11):4130-5. doi: 10.1128/AAC.48.11.4130-4135.2004. Antimicrob Agents Chemother. 2004. PMID: 15504831 Free PMC article.
References
-
- Barrineau P, Gilbert M P, Jackson W J, Jones C S, Summers A O, Wisdom S. The DNA sequence of the mercury resistance operon of the IncFII plasmid NR1. J Mol Appl Genet. 1984;2:601–619. - PubMed
-
- Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

