Gliding motility in bacteria: insights from studies of Myxococcus xanthus
- PMID: 10477310
- PMCID: PMC103748
- DOI: 10.1128/MMBR.63.3.621-641.1999
Gliding motility in bacteria: insights from studies of Myxococcus xanthus
Abstract
Gliding motility is observed in a large variety of phylogenetically unrelated bacteria. Gliding provides a means for microbes to travel in environments with a low water content, such as might be found in biofilms, microbial mats, and soil. Gliding is defined as the movement of a cell on a surface in the direction of the long axis of the cell. Because this definition is operational and not mechanistic, the underlying molecular motor(s) may be quite different in diverse microbes. In fact, studies on the gliding bacterium Myxococcus xanthus suggest that two independent gliding machineries, encoded by two multigene systems, operate in this microorganism. One machinery, which allows individual cells to glide on a surface, independent of whether the cells are moving alone or in groups, requires the function of the genes of the A-motility system. More than 37 A-motility genes are known to be required for this form of movement. Depending on an additional phenotype, these genes are divided into two subclasses, the agl and cgl genes. Videomicroscopic studies on gliding movement, as well as ultrastructural observations of two myxobacteria, suggest that the A-system motor may consist of multiple single motor elements that are arrayed along the entire cell body. Each motor element is proposed to be localized to the periplasmic space and to be anchored to the peptidoglycan layer. The force to glide which may be generated here is coupled to adhesion sites that move freely in the outer membrane. These adhesion sites provide a specific contact with the substratum. Based on single-cell observations, similar models have been proposed to operate in the unrelated gliding bacteria Flavobacterium johnsoniae (formerly Cytophaga johnsonae), Cytophaga strain U67, and Flexibacter polymorphus (a filamentous glider). Although this model has not been verified experimentally, M. xanthus seems to be the ideal organism with which to test it, given the genetic tools available. The second gliding motor in M. xanthus controls cell movement in groups (S-motility system). It is dependent on functional type IV pili and is operative only when cells are in close proximity to each other. Type IV pili are known to be involved in another mode of bacterial surface translocation, called twitching motility. S-motility may well represent a variation of twitching motility in M. xanthus. However, twitching differs from gliding since it involves cell movements that are jerky and abrupt and that lack the organization and smoothness observed in gliding. Components of this motor are encoded by genes of the S-system, which appear to be homologs of genes involved in the biosynthesis, assembly, and function of type IV pili in Pseudomonas aeruginosa and Neisseria gonorrhoeae. How type IV pili generate force in S-motility is currently unknown, but it is to be expected that ongoing physiological, genetic, and biochemical studies in M. xanthus, in conjunction with studies on twitching in P. aeruginosa and N. gonorrhoeae, will provide important insights into this microbial motor. The two motility systems of M. xanthus are affected to different degrees by the MglA protein, which shows similarity to a small GTPase. Bacterial chemotaxis-like sensory transduction systems control gliding motility in M. xanthus. The frz genes appear to regulate gliding movement of individual cells and movement by the S-motility system, suggesting that the two motors found in this bacterium can be regulated to result in coordinated multicellular movements. In contrast, the dif genes affect only S-system-dependent swarming.
Figures


 •
• ▸).
(B) In this model, two types of unidirectional motors exist
(•
▸).
(B) In this model, two types of unidirectional motors exist
(• ▸ and
◂
▸ and
◂ •), each capable of generating
force in only one direction. The motor elements are arranged opposite
each other, so that selected activation of one or the other results in
movement in one or the other direction.
•), each capable of generating
force in only one direction. The motor elements are arranged opposite
each other, so that selected activation of one or the other results in
movement in one or the other direction.



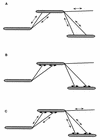
 ○
○ ▸),
e.g., which are part of the gliding motor of A motility and are
involved in A-motility-dependent gliding. Linked to the moving adhesion
sites by a pilus, a neighboring cell is “dragged” along, thus
performing a translocation in the direction of the long axis of the
cell. (C) A combination of pilus retraction-extension and adhesion
site-dependent displacement (A plus B).
▸),
e.g., which are part of the gliding motor of A motility and are
involved in A-motility-dependent gliding. Linked to the moving adhesion
sites by a pilus, a neighboring cell is “dragged” along, thus
performing a translocation in the direction of the long axis of the
cell. (C) A combination of pilus retraction-extension and adhesion
site-dependent displacement (A plus B).
References
-
- Abbant D R, Leadbetter E R, Godchaux W I, Escher A. Sulfonolipids are molecular determinants of gliding motility. Nature. 1986;324:367–369.
-
- Alm R A, Mattick J S. Genes involved in the biogenesis and function of type-4 fimbriae in Pseudomonas aeruginosa. Gene. 1997;192:89–98. - PubMed
-
- Armitage J P, Schmitt R. Bacterial chemotaxis: Rhodobacter sphaeroides and Sinorhizobium meliloti: variations on a theme. Microbiology. 1997;143:3671–3682. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

