Metabolism and genetics of Helicobacter pylori: the genome era
- PMID: 10477311
- PMCID: PMC103749
- DOI: 10.1128/MMBR.63.3.642-674.1999
Metabolism and genetics of Helicobacter pylori: the genome era
Abstract
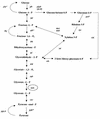
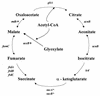
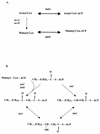
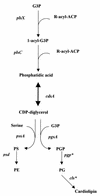
The publication of the complete sequence of Helicobacter pylori 26695 in 1997 and more recently that of strain J99 has provided new insight into the biology of this organism. In this review, we attempt to analyze and interpret the information provided by sequence annotations and to compare these data with those provided by experimental analyses. After a brief description of the general features of the genomes of the two sequenced strains, the principal metabolic pathways are analyzed. In particular, the enzymes encoded by H. pylori involved in fermentative and oxidative metabolism, lipopolysaccharide biosynthesis, nucleotide biosynthesis, aerobic and anaerobic respiration, and iron and nitrogen assimilation are described, and the areas of controversy between the experimental data and those provided by the sequence annotation are discussed. The role of urease, particularly in pH homeostasis, and other specialized mechanisms developed by the bacterium to maintain its internal pH are also considered. The replicational, transcriptional, and translational apparatuses are reviewed, as is the regulatory network. The numerous findings on the metabolism of the bacteria and the paucity of gene expression regulation systems are indicative of the high level of adaptation to the human gastric environment. Arguments in favor of the diversity of H. pylori and molecular data reflecting possible mechanisms involved in this diversity are presented. Finally, we compare the numerous experimental data on the colonization factors and those provided from the genome sequence annotation, in particular for genes involved in motility and adherence of the bacterium to the gastric tissue.
Figures


Similar articles
-
The physiology and metabolism of the human gastric pathogen Helicobacter pylori.Adv Microb Physiol. 1998;40:137-89. doi: 10.1016/s0065-2911(08)60131-9. Adv Microb Physiol. 1998. PMID: 9889978 Review.
-
Helicobacter pylori physiology predicted from genomic comparison of two strains.Microbiol Mol Biol Rev. 1999 Sep;63(3):675-707. doi: 10.1128/MMBR.63.3.675-707.1999. Microbiol Mol Biol Rev. 1999. PMID: 10477312 Free PMC article. Review.
-
Analysis of the genetic diversity of Helicobacter pylori: the tale of two genomes.J Mol Med (Berl). 1999 Dec;77(12):834-46. doi: 10.1007/s001099900067. J Mol Med (Berl). 1999. PMID: 10682319 Review.
-
The Helicobacter pylori Methylome: Roles in Gene Regulation and Virulence.Curr Top Microbiol Immunol. 2017;400:105-127. doi: 10.1007/978-3-319-50520-6_5. Curr Top Microbiol Immunol. 2017. PMID: 28124151 Review.
-
Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori.Mol Microbiol. 2004 Jul;53(2):623-38. doi: 10.1111/j.1365-2958.2004.04137.x. Mol Microbiol. 2004. PMID: 15228539
Cited by
-
Oxidative-stress resistance mutants of Helicobacter pylori.J Bacteriol. 2002 Jun;184(12):3186-93. doi: 10.1128/JB.184.12.3186-3193.2002. J Bacteriol. 2002. PMID: 12029034 Free PMC article.
-
Gene expression profiling of Helicobacter pylori reveals a growth-phase-dependent switch in virulence gene expression.Infect Immun. 2003 May;71(5):2643-55. doi: 10.1128/IAI.71.5.2643-2655.2003. Infect Immun. 2003. PMID: 12704139 Free PMC article.
-
Helicobacter urease: niche construction at the single molecule level.J Biosci. 2009 Oct;34(4):503-11. doi: 10.1007/s12038-009-0069-4. J Biosci. 2009. PMID: 19920336
-
Complexomics study of two Helicobacter pylori strains of two pathological origins: potential targets for vaccine development and new insight in bacteria metabolism.Mol Cell Proteomics. 2010 Dec;9(12):2796-826. doi: 10.1074/mcp.M110.001065. Epub 2010 Jul 7. Mol Cell Proteomics. 2010. PMID: 20610778 Free PMC article.
-
Vitamin B6 is required for full motility and virulence in Helicobacter pylori.mBio. 2010 Aug 17;1(3):e00112-10. doi: 10.1128/mBio.00112-10. mBio. 2010. PMID: 21151756 Free PMC article.
References
-
- Alderson J, Clayton C L, Kelly D J. Investigations into the aerobic respiratory chain of Helicobacter pylori. Gut. 1997;41:A7. . (Abstract.)
-
- Alm R A, Ling L-S L, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, de Jonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. - PubMed
-
- Apel I, Jacobs E, Kist M, Bredt W. Antibody response of patients against a 120 kDa surface protein of Campylobacter pylori. Zentbl Bakteriol Mikrobiol Hyg Ser A. 1988;268:271–276. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

